The projects in this repository demonstrate working with genomic data via Google BigQuery. All examples are built upon public datasets.
You can execute these examples by:
- Copying and pasting the queries into
- the BigQuery Browser Tool
- the bq Command-Line Tool
- one of the many third-party tools that have integrated BigQuery
- Running the chunks of R code within the RMarkdown files in R or RStudio
- Running the chunks of Python code within the iPython Notebooks in iPython
With minor modification, you can run the same analyses on your own genomic data within BigQuery.
- Set up a BigQuery project.
- Follow the BigQuery instructions on how to sign up for BigQuery and set up a new API project.
- You can use the Team tab to share a project with co-workers and colleagues.
- Run a query.
- go to the BigQuery Browser Tool
- click on the Compose query button
- paste the SQL query for 1,000 Genomes indel length counts into the query textbox
- click Run query to get your results
- Note: you do not need to enable billing to run the smaller queries.
- All of the queries for the small phenotypic dataset available in the data story "Exploring the phenotypic data" should be runnable in this free mode.
- If you see an
Exceeded quotaerror, that means you will need to enable billing and you will be charged for that query. See BigQuery pricing for more detail. - For example, queries within the 1,000 Genomes dataset that examine sample genotype columns will process approximately 1TB of data per query. (1,000 GB * $0.005 per GB processed = $5.00)
- Add the public datasets to your project so that they show up in the left-hand naviation pane
- go to the BigQuery Browser Tool
- click on the drop down icon beside your project name in the left navigator
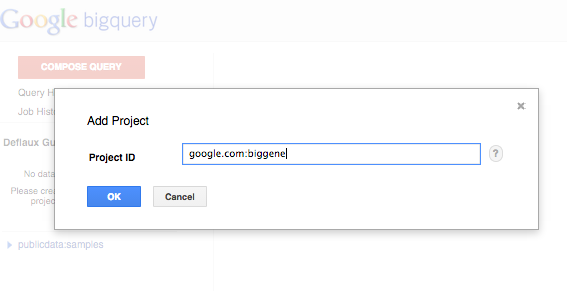
- pick ‘Switch to project’ in the menu, and ‘Display project...’ in the submenu
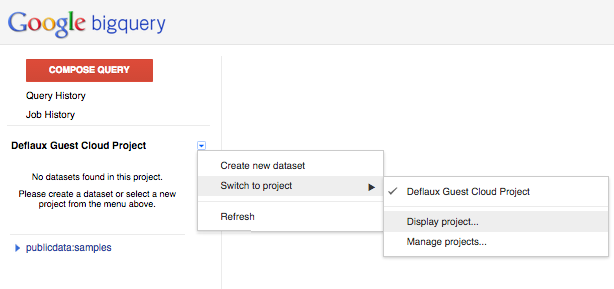
- enter google.com:biggene in the ‘Add Project’ dialog
- What next?
- New to BigQuery? See the query reference.
- New to working with variants? See an overview of the VCF data format.
Sample analyses upon VCF data from the 1,000 Genomes Project
Project Name: google.com:biggene
Sample analyses upon the Personal Genome Project
Project Name: google.com:biggene
The Google Genomics API spec includes a not-yet-implemented import method that loads VCF files directly from Cloud Storage. Until an implementation of the method is available, you will need to transform your VCF data into JSON with a schema similar to what you see in these examples, and then load the JSON into BigQuery. See Preparing Data for BigQuery and also BigQuery in Practice : Loading Data Sets That are Terabytes and Beyond for more detail.
The Google Genomics Discuss mailing list is a good
way to sync up with other people who use googlegenomics including the core developers. You can subscribe
by sending an email to google-genomics-discuss+subscribe@googlegroups.com or just post using
the web forum page.