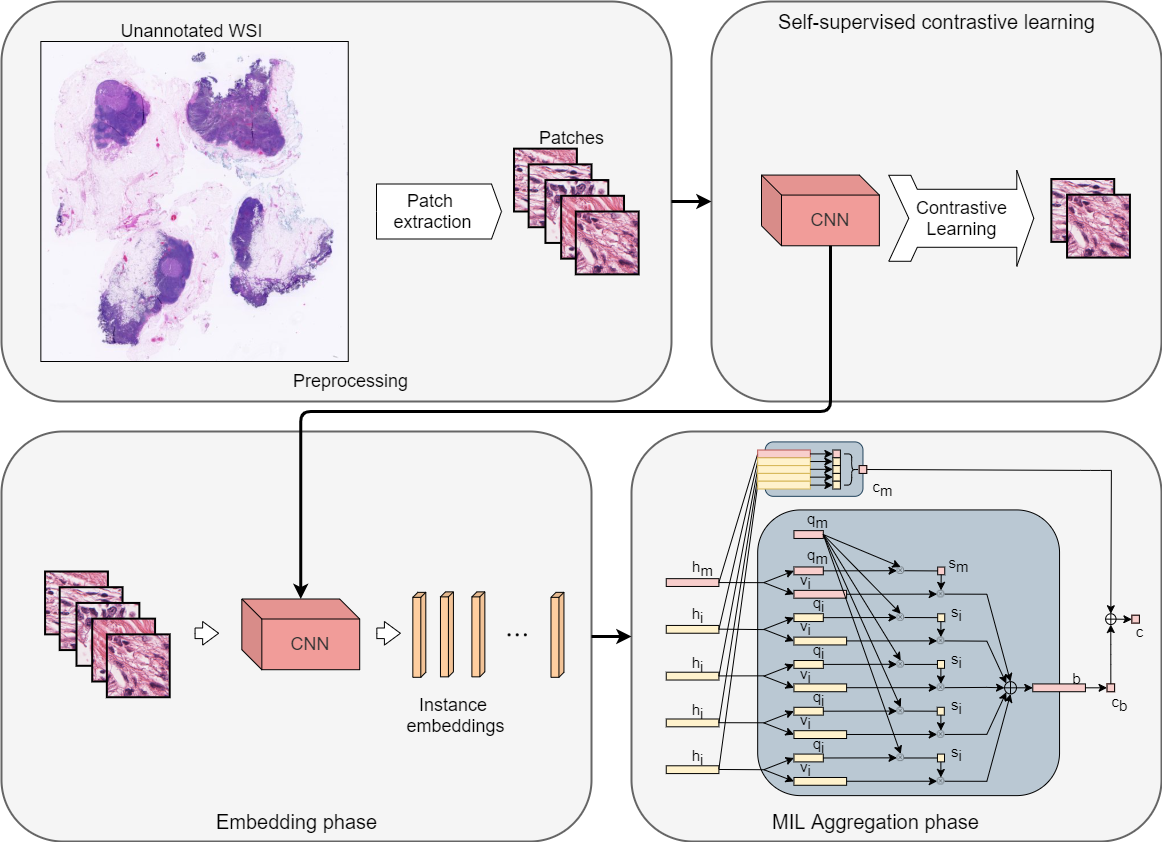
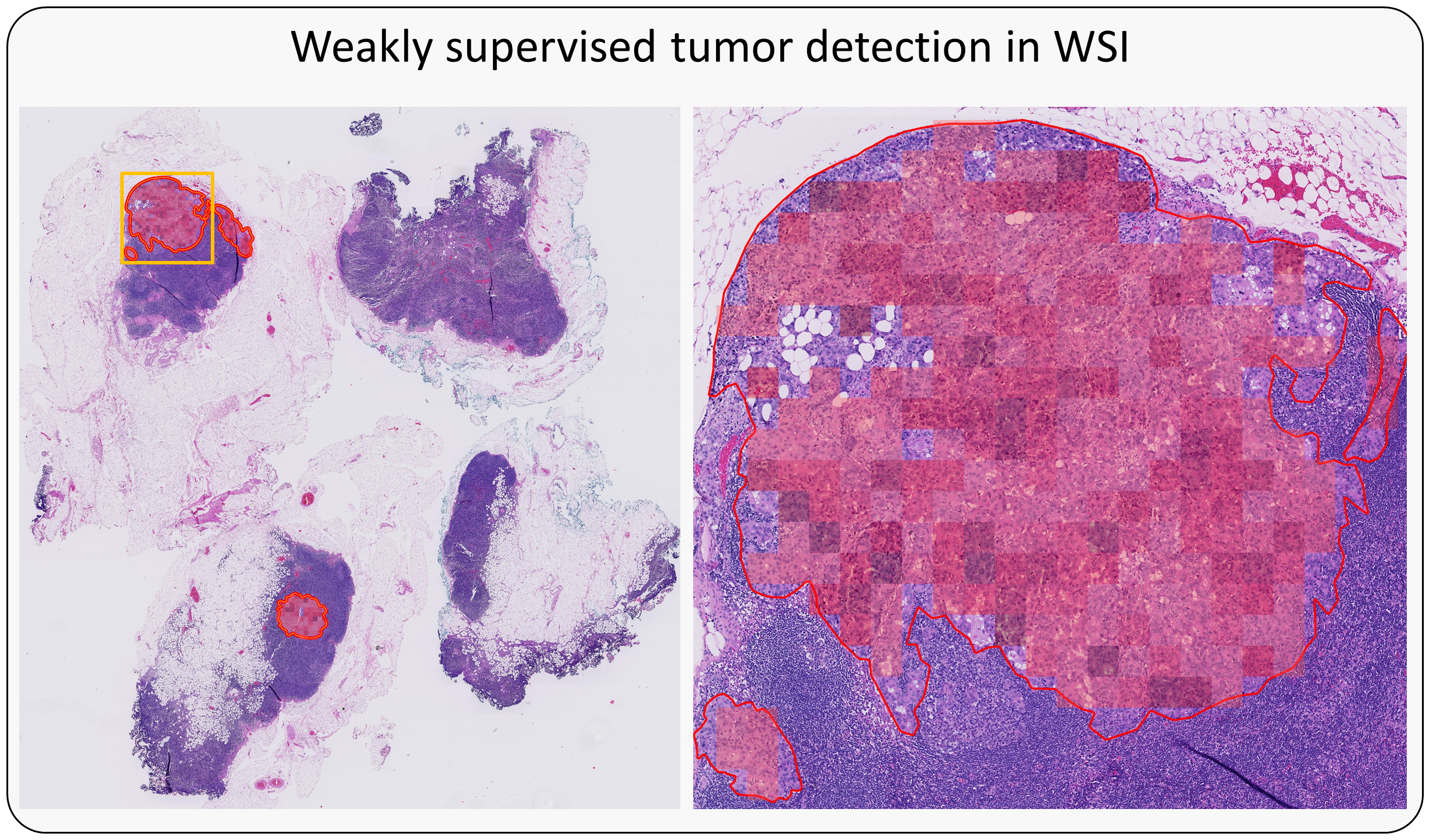
This is the Pytorch implementation for the multiple instance learning model described in the paper Dual-stream Multiple Instance Learning Network for Whole Slide Image Classification with Self-supervised Contrastive Learning (CVPR 2021, accepted for oral presentation).
Install anaconda/miniconda
Required packages
$ conda env create --name dsmil --file env.yml
$ conda activate dsmil
Install OpenSlide and openslide-python.
Tutorial 1 and Tutorial 2 (Windows).
MIL benchmark datasets can be downloaded via:
$ python download.py --dataset=mil
Precomputed features for TCGA Lung Cancer dataset can be downloaded via:
$ python download.py --dataset=tcga
This dataset requires 20GB of free disk space.
Train DSMIL on standard MIL benchmark dataset:
$ python train_mil.py
Switch between MIL benchmark dataset, use option:
[--datasets] # musk1, musk2, elephant, fox, tiger
Other options are available for learning rate (--lr=0.0002), cross validation fold (--cv_fold=10), weight-decay (--weight_decay=5e-3), and number of epochs (--num_epoch=40).
Train DSMIL on TCGA Lung Cancer dataset (precomputed features):
$ python train_tcga.py --new_features=0
We provided a testing pipeline for several sample slides. The slides can be downloaded via:
$ python download.py --dataset=tcga-test
To crop the WSIs into patches, run:
$ python TCGA_test_10x.py
A folder containing all patches for each WSI will be created at
./test/patches.
After the WSIs are cropped, run the testing script:
$ python testing.py
The thumbnails of the WSIs will be saved in
./test/thumbnails.
The detection color maps will be saved in./test/output.
The testing pipeline will process every WSI placed inside the./test/inputfolder. The slide will be detected as a LUAD, LUSC, or benign sample.
If you are processing WSI from raw images, you will need to download the WSIs first.
Download WSIs.
From GDC data portal. You can use GDC data portal with a manifest file and configuration file. Example manifest files and configuration files can be found in tcga-download. The raw WSIs take about 1TB of disc space and may take several days to download. Open command-line tool (Command Prompt for the case of Windows), unzip the data portal client into
./tcga-download, navigate to./tcga-download, and use commands:
$ cd tcga-download
$ gdc-client -m gdc_manifest.2020-09-06-TCGA-LUAD.txt --config config-LUAD.dtt
$ gdc-client -m gdc_manifest.2020-09-06-TCGA-LUSC.txt --config config-LUSC.dtt
The data will be saved in
./WSI/TCGA-lung. Please check details regarding the use of TCGA data portal. Otherwise, individual WSIs can be download manually in GDC data portal repository
From Google Drive. The svs files are also uploaded. The dataset contains in total 1053 slides, including 512 LUSC and 541 LUAD. 10 low-quality LUAD slides are discarded.
Prepare the patches.
We will be using OpenSlide, a C library with a Python API that provides a simple interface to read WSI data. We refer the users to OpenSlide Python API document for the details of using this tool.
The patches could be saved in './WSI/TCGA-lung/pyramid' in a pyramidal structure for the magnifications of 20x and 5x. Run:
$ python WSI_cropping.py --multiscale=1
Or, the patches could be cropped at a single magnification of 10x and saved in './WSI/TCGA-lung/single' by using:
$ python WSI_cropping.py --multiscale=0
Train the embedder.
We provided a modified script from this repository Pytorch implementation of SimCLR For training the embedder.
Navigate to './simclr' and edit the attributes in the configuration file 'config.yaml'. You will need to determine a batch size that fits your gpu(s). We recommend using a batch size of at least 512 to get good simclr features. The trained model weights and loss log are saved in folder './simclr/runs'.
cd simclr
$ python run.py
Compute the features.
Compute the features for 20x magnification:
$ cd ..
$ python compute_feats.py --dataset=wsi-tcga-lung
Or, compute the features for 10x magnification:
$ python compute_feats.py --dataset=wsi-tcga-lung-single --magnification=10x
Start training.
$ python train_tcga.py --new_features=1
- Place WSI files into
WSI\[DATASET_NAME]\[CATEGORY_NAME]\[SLIDE_FOLDER_NAME] (optional)\SLIDE_NAME.svs.
For binary classifier, the negative class should have
[CATEGORY_NAME]at index0when sorted alphabetically. For multi-class classifier, if you have a negative class (not belonging to any of the positive classes), the folder should have[CATEGORY_NAME]at the last index when sorted alphabetically. The naming does not matter if you do not have a negative class.
- Crop patches.
$ python WSI_cropping.py --dataset=[DATASET_NAME]
- Train an embedder.
$ cd simclr
$ python run.py --dataset=[DATASET_NAME]
- Compute features using the embedder.
$ cd ..
$ python compute_feats.py --dataset=[DATASET_NAME]
This will use the last trained embedder to compute the features, if you want to use an embedder from a specific run, add the option
--weights=[RUN_NAME], where[RUN_NAME]is a folder name insidesimclr/runs/. If you have an embedder you want to use, you can place the weight file assimclr/runs/[FOLDER_NAME]/checkpoints/model.pthand pass the[FOLDER_NAME]to this option. The embedder architecture is ResNet18.
- Training.
$ python train_tcga.py --dataset=[DATASET_NAME] --new_features=1
You will need to adjust
--num_classesoption if the dataset contains more than 2 positive classes or only 1 positive class. See the next section for details.
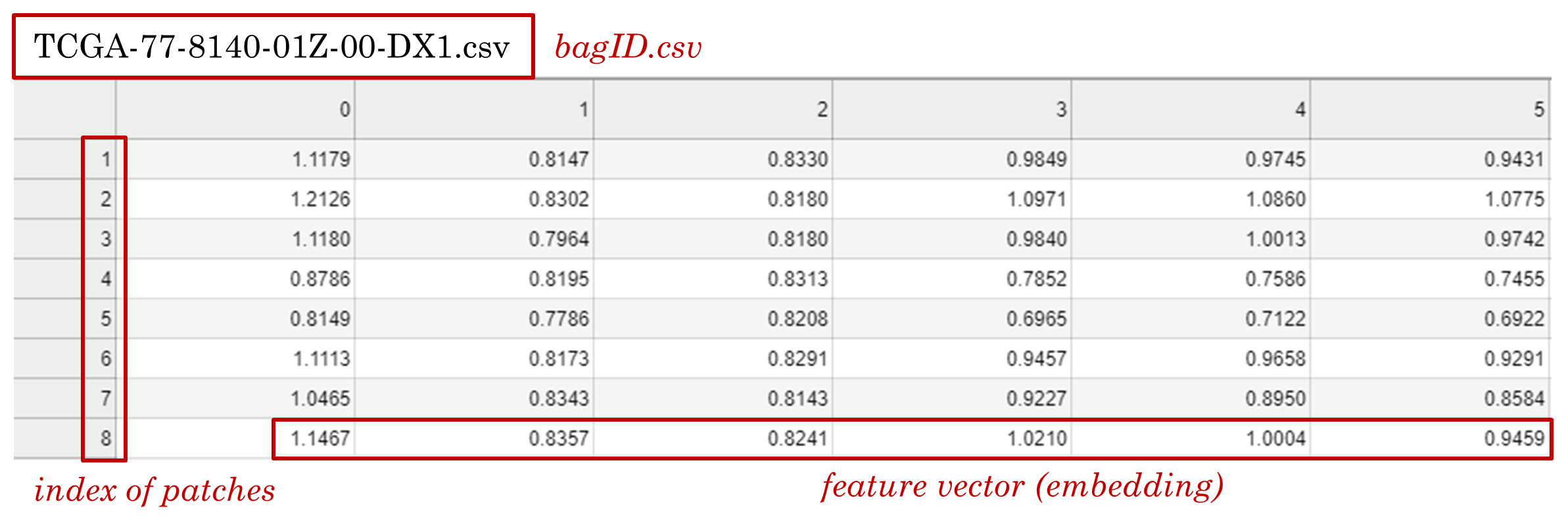
- For each bag, generate a .csv file where each row contains the feature of an instance. The .csv file should be named as "bagID.csv" and put into a folder named "dataset-name/category/".
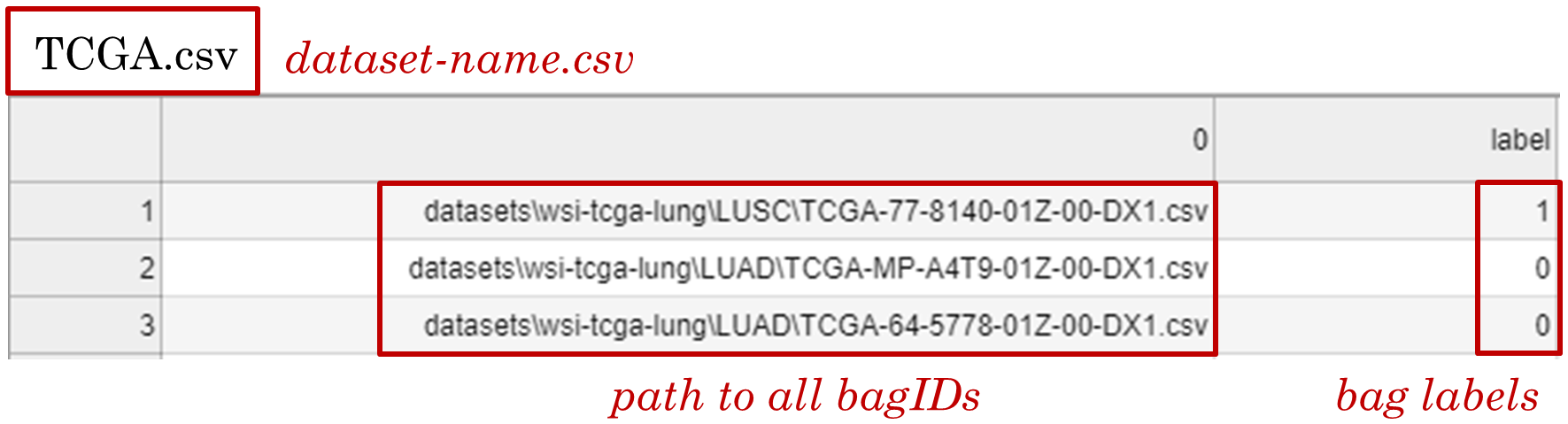
- Generate a "dataset-name.csv" file with two columns where the first column contains the paths to all bagID.csv files, and the second column contains the bag labels.
- Labels.
For binary classifier, use
1for positive bags and0for negative bags. Use--num_classes=1at training.
For multi-class classifier (Npositive classes and one optional negative class), use0~(N-1)for positive classes. If you have negative class (not belonging to any one of the positive classes), useNfor its label. Use--num_classes=N(Nequals the number of positive classes) at training.
If you use the code or results in your research, please use the following BibTeX entry.
@article{li2020dual,
title={Dual-stream Multiple Instance Learning Network for Whole Slide Image Classification with Self-supervised Contrastive Learning},
author={Li, Bin and Li, Yin and Eliceiri, Kevin W},
journal={arXiv preprint arXiv:2011.08939},
year={2020}
}