A command line tool for a pangenome analysis from proteinotho calculated cluster table and description files using python 3.8. Add gene descriptions to the tabular output of cluster tables from proteinortho calculations. Retrieve a tabular pangenome analysis of various proteinortho cluster tables of from one calculation. So now you can compare the various cluster output tables from proteinorto.
This is a command line tool using python version 3.8. So make sure to have python version 3.8 installed and use it as python interpreter.
To get the usage information run:
python main.py -h
This will display the following information:
usage: main.py [-h] [-i INPUTFILE [INPUTFILE ...]] [-d DESCRIPTION] [-N] [-A] [-V] [-G] [-v]
optional arguments:
-h, --help show this help message and exit
-i INPUTFILE [INPUTFILE ...], --inputfile INPUTFILE [INPUTFILE ...]
path to tabular proteinortho output file(s) required for options -A/-G/-N/-V
-d DESCRIPTION, --description DESCRIPTION
path to tabular proteinortho description file
-N, --NAMES add gene names to IDs for the tabular input file
-A, --ANALYSE tabular pangenome analysis
-V, --VISUALIZE visualization of the pangenome composition as pie or bar plot
-G, --GENE_CONTRIBUTION
box plot of gene contribution with each genome
-v, --version show program version
1. -A/--ANALYSE
Here is an example for running the -A/--ANALYSE-option with the cluster table form proteinortho's calculation including 5 available genomes of the genus Saccharospirillum. The second input table was retrieved by running the program with the -N/--NAMES-option.
Command:
python main.py \
-d Saccharospirillum.descriptions \
-i Saccharospirillum.poff.tsv Saccharospirillum.poff.named.tsv \
-A
Output:
test/Saccharospirillum.poff.tsv test/Saccharospirillum.poff.named.tsv
Total genes from all input genomes 19444 19444
Genes in gene clusters 17267 17267
Gene clusters 4379 4379
Unique genes 2177 2177
Core gene clusters 2344 2344
Accessory gene clusters 2035 2035
Core genes in clusters 11730 11730
Accessory genes in clusters 5537 5537
Gene cluster with annotated function 0 2566
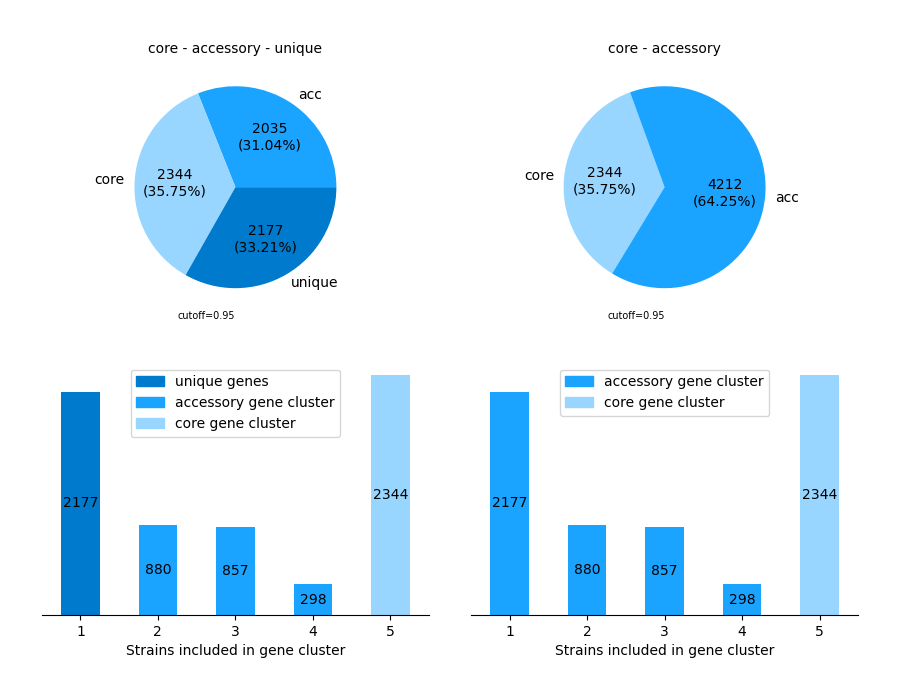
2. -V/--VISUALIZE
Get an overview of the pangenome by choosing the -V/--VISUALIZE-option, retrieving two pie charts and two bar charts.
Command:
python main.py \
-d Saccharospirillum.descriptions \
-i Saccharospirillum.poff.tsv \
-V
Output:
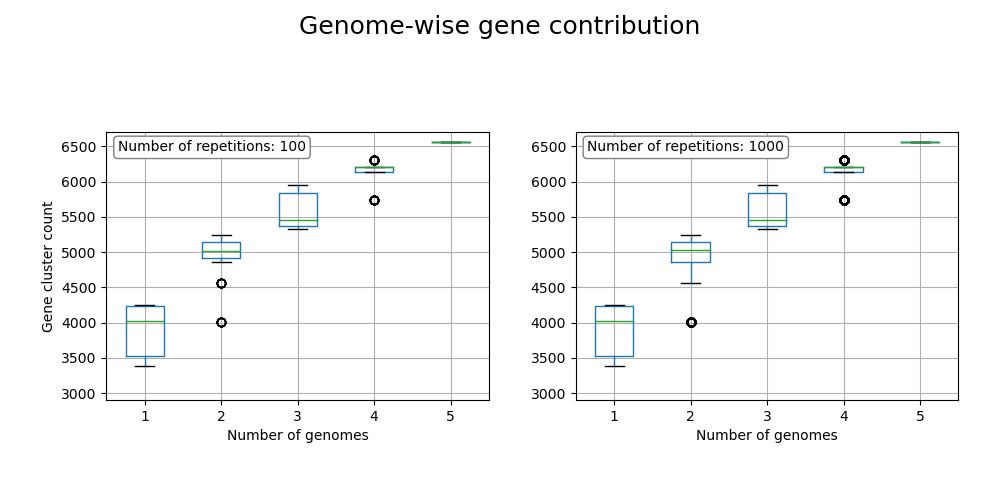
3. -G/--GENE_CONTRIBUTION
The gene contribution with each genome to the pangenome is calcualted in several iterations. If at least the lines in the last two columns are at same height, no gene contribution is happening with adding a further genome, therefore the pangenome is closed. Otherwise, the pangenome is open.
Command:
python main.py \
-d Saccharospirillum.descriptions \
-i Saccharospirillum.poff.tsv \
-G
Output: