The content in this repository is for the workshop on reproducibility in R, given at the Western Society of Naturalists meeting, November 09, 2023.
Start by selecting somewhere on your computer for this directory to live. It usually make sense to keep all your folders for different projects in the same place. So we're all on the same page, we suggest naming it wsn-reproducibility. Now, we'll be making some folders to keep our files in. We'll make four folders to start:
/data/raw/clean
/R/outputs/figs
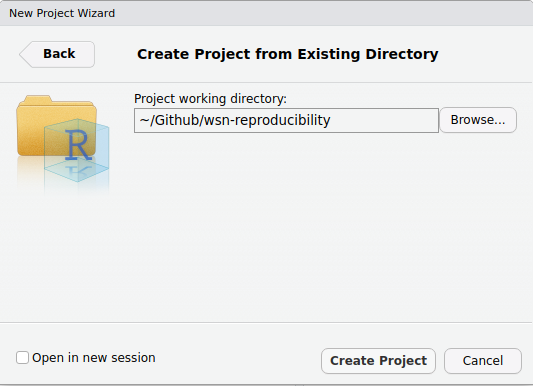
These three are the basis of most analysis projects. Next, we'll make an RProject. To do this, open RStudio, then click File > New Project > Existing Directory, then use the Browse... button to navigate in your folder structure and select the wsn-reproducibility folder.
We're also going to be making a file in the root of our directory calle __main.R. This will be the file that we use to operationalize our "one-click" principle, where we can simply re-run an entire analysis, maybe even with new data, by clicking a single button.
In this workshop, we're going to be using data that will be pulled from a Google Drive location. To do this, we'll need to write some code in some R files. Start my making a new file in the /R folder called 00_data_wrangling.R.
In this file will be where we read in the data from the web, and clean it.
In this workshop, because of how we're showing you how to set up a working set up, we're going to be using explicit package specification for every single function. This is for two reasons:
- We don't want to have to call our packages more than once (this is more efficient), so the package calls will be in the
__main.Rfile. - Pedagogically, it makes it more clear which packages are involved.
Okay, back to data from the web. In our file, start with some basic info at the top of the file including your name, the date, and what this is supposed to be doing in the file (in a few words). In our workflow of writing code, remember we're going to be continuously updating our4 __main.R file with the packages you're using and the files we need.
For the code we need in this file, refer to the file directly here in the repo. Here are the steps in normal people speak:
- Define the URL and download the file
- Read in the file
- Remove any NA's from the species richness file
- Rename the columns
- Write both out to a
/data/clean/location
Because we're limited on time, we'll be doing only part of the modeling process. We'll only be comparing two models and not going through all of the diagnostic checking. That is left as an exercise for those interested. What we'll do is:
- Read in our cleaned data
- Check the data structures
- Fit two competing models
- Use AIC as our metric to compare the models
- Write out the best model
We now have our final model, great! However, we need to come up with a way to display our results. A commmon way of doing this is to plot the
- Note you could just pull the data directly into a dataframe with read_csv() but we aren't doing that here for demonstration purposes and because it gives you a copy in case you want to work offline which for field folks is often
- Look at the column types then build out the ifelse
- Talk through the relationship between species richness and biomass very briefly
- Link functions are discussed in the tutorial for those who need a refresher
- For the AIC, do AIC(mod1, mod2), then break it into two for the ifelse
- save the best model that we've named separately
- Go through it step by step
- Remember each function needs a forcing