Convert AlphaFold distograms into distance matrices and save them into a number of formats.
The distances for a residue pair
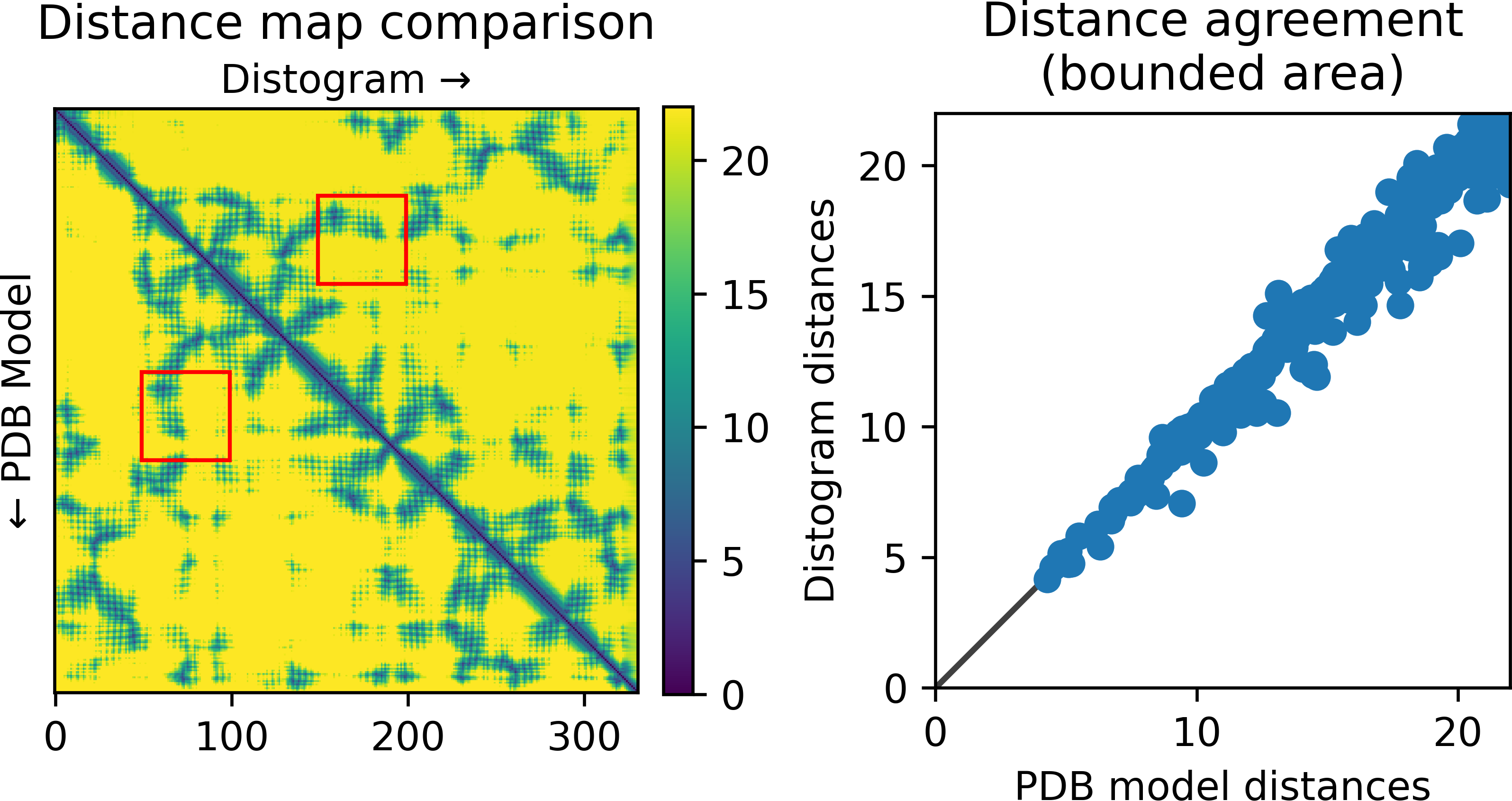
Distances calculated this way agree quite well with the actual
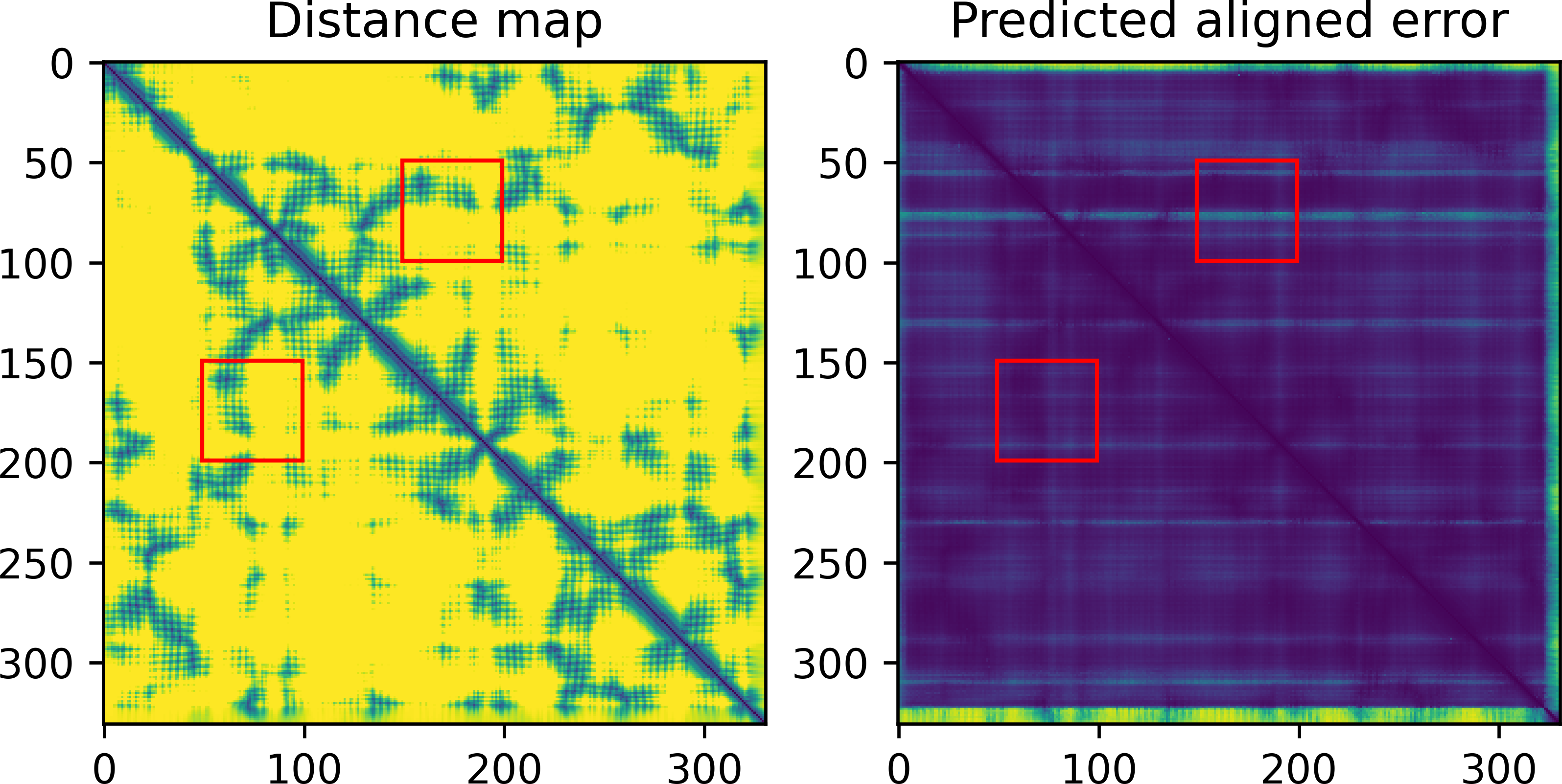
dgram2dmap allows to plot the converted distance maps and compare them with the predicted aligned error, while highlighting distances between numbered subsets of amino acids or chain identifiers:
It also allows to compare against a 3D model:
usage: dgram2dmap.py [-h] [--maxD 20.0] [--limits i:j k:l] [--chains chain1 chain2] [--plot] [--argmax] [--rosetta] [--pdb ranked_0.pdb] in_folder
Extract and format distance constraints from AlphaFold distograms
positional arguments:
in_folder AlphaFold model output folder
optional arguments:
-h, --help show this help message and exit
--maxD 20.0 Maximum distance (in Å) for constraints output
--limits i:j k:l Select a 'patch' of constraints between two subsets of residues (e.g. 0:100 200:300)
--chains chain1 chain2
Extract constraints between two chains (e.g. A B)
--plot Plot the distances with bounding boxes
--argmax Use argmax to find the most likely distance instead of interpolating
--rosetta Export below-threshold (see maxD) distances in a Rosetta constraint files
--pdb model.pdb PDB model of the target protein (e.g. for comparisons against native)
Example:
If you want to extract the distances from the pickle files for CASP15 target T1105 (available here) while extracting the distances between residues 50:100 and 150:200 to a rosetta constraint file:
python dgram_to_dmap.py AF_outputs/T1105/ --chains 50:100 150:200 --plot --rosetta
Which will produce the following outputs for each pickle file (see also example/ folder):
result_model_1_ptm_pred_0.pkl.rosetta_constraints: rosetta constraint file from selected rangesresult_model_1_ptm_pred_0.pkl.dmap: CSV file with all distances calculated from the distogramresult_model_1_ptm_pred_0.pkl.dmap.png: image of the calculated distances (with selection range boxes, if any). The predicted aligned error is also shown if present:result_model_1_ptm_pred_0.pkl.agreement.png: comparison between model (lower corner) and distogram (upper corner) distance map, scatter plot of model vs. distogram distances in range boxes (see plot above)