Implementation of several predictive coding models
Gagnepain, P., Henson, R. N., & Davis, M. H. (2012). Temporal predictive codes for spoken words in auditory cortex. Current Biology, 22(7), 615-621.
-
Go to
Gagnepain2012and run 'ipython'cd Gagnepain2012 ipython -
Import model
from Gagnepain2012 import Model -
Generate model
new_Model = Model( lexicon_File= <path>, additional_Lexicon_File= <path> )- Parameters
lexicon_File- The lexicon that the model uses by default
- 'ELP_groupData.csv' is an example
additional_Lexicon_File- Lexicon assuming further learning
- 'Novel_Lexicon.csv' is an example
- Parameters
-
Test
new_Model.Test(phoneme_String= <str>)- Result example
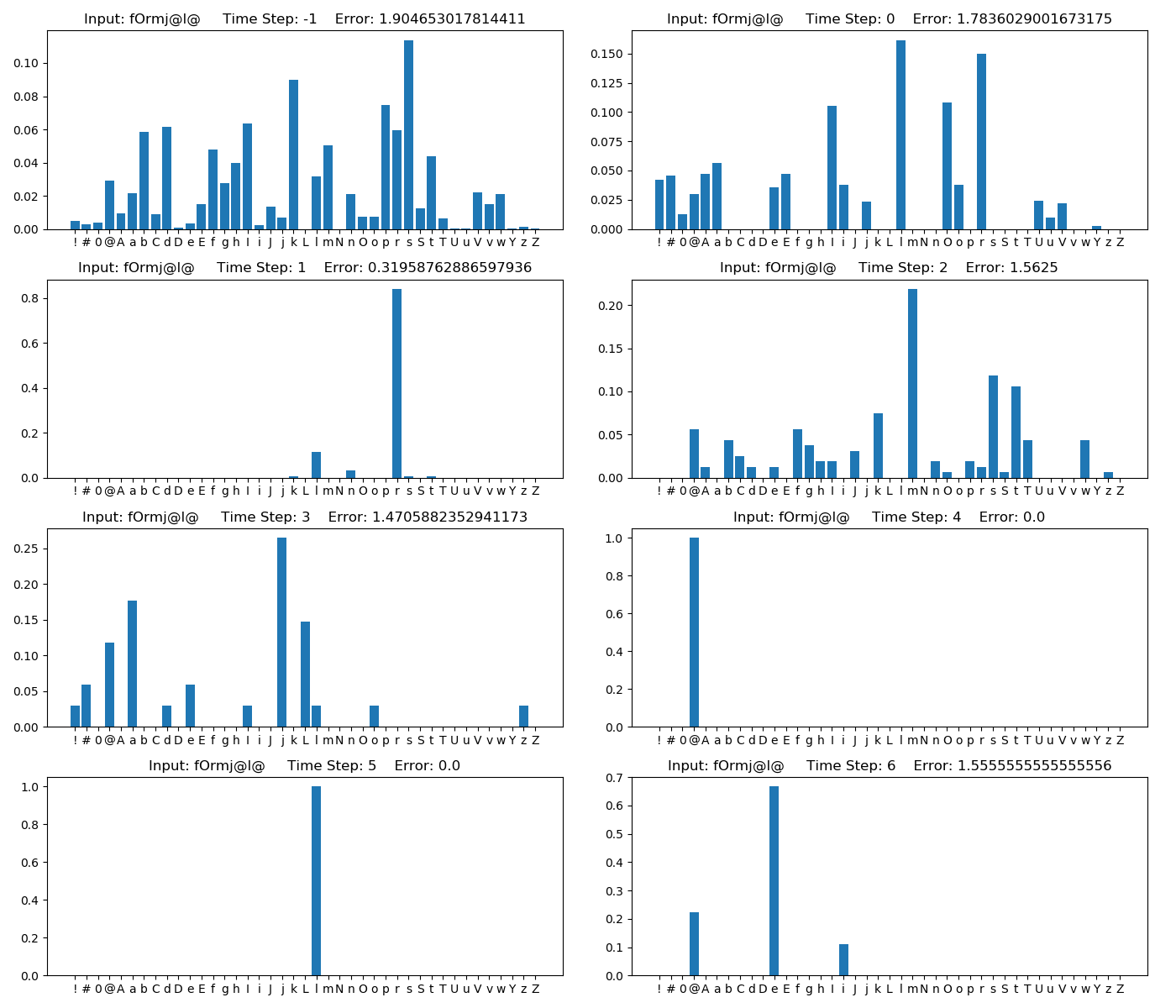
- We made a jupyter notebook example to use SRN model. Please check Train.ipynb and Test.ipynb
-
Go to
SRNand run 'ipython'cd SRN ipython -
Import model
from SRN import Model, Sigmoid, Softmax, List_Test -
Generate model
new_Model = Model( hidden_Unit= <int>, output_Function= <Sigmoid or Softmax>, lexicon_File= <path>, additional_Lexicon_File= <path>, weight_File= <path>, use_Frequency= <bool> )- Parameters
hidden_Unit- The size of hidden units
output_Function- Determine output function
SoftmaxorSigmoid
lexicon_File- The lexicon that the model uses by default
ELP_groupData.csvis an example
additional_Lexicon_File- Lexicon assuming further learning
Novel_Lexicon_1.csvandNovel_Lexicon_2.csvare examples.
weight_File- If you want load a pre-trained weight file, set the weight path
- If not, set
None
use_Frequency- If you want to use frequency information of lexicon, set
True - Default is
False
- If you want to use frequency information of lexicon, set
- Parameters
-
Train basic lexicon
new_Model.Train( learning_Rate= <float>, max_Epoch= <int>, epoch_Batch_Size= <int>, mode='Normal', test_Pronunciation = <str> )- Parameters
learning_Rate- The learning rate while training.
max_Epoch- Determine the maximum training epoch.
epoch_Batch_Size- Determine the batch size of training.
- After doing batch training, the weight will be saved.
mode- In basic lexicon training, this parameter is fixed 'Normal'
test_Pronunciation- Determine one phoneme string will be tested While training.
- Parameters
-
Train additional lexicon
new_Model.Train( learning_Rate= <float>, max_Epoch= <int>, epoch_Batch_Size= <int>, mode='Addition', test_Pronunciation = <str>, initial_Epoch= <int>, tag= <str>, )- Parameters
learning_Rate- The learning rate while training.
- Using lower value than basic lexicon training's is recommended.
max_Epoch- Determine the maximum training epoch.
epoch_Batch_Size- Determine the batch size of training.
- After doing batch training, the weight will be saved.
mode- In basic lexicon training, this parameter is fixed 'Addition'
test_Pronunciation- Determine a phoneme string will be tested While training.
initial_Epoch- Set number the initial epoch.
- This value does not affect the model's performance.
- Using the last epoch of
Pretraining is recommended to manage easily.
- Parameters
-
Single phoneme string test
-
Load both of two pre and post addition models.
pre_Model = Model( hidden_Unit= <int>, output_Function= <Sigmoid or Softmax>, lexicon_File= <path>, additional_Lexicon_File= <path>, weight_File= <path> ) post_Model = Model( hidden_Unit= <int>, output_Function= <Sigmoid or Softmax>, lexicon_File= <path>, additional_Lexicon_File= <path>, weight_File= <path> )weight_Fileis located in './Results/<Use_Frequency>/Weight'
-
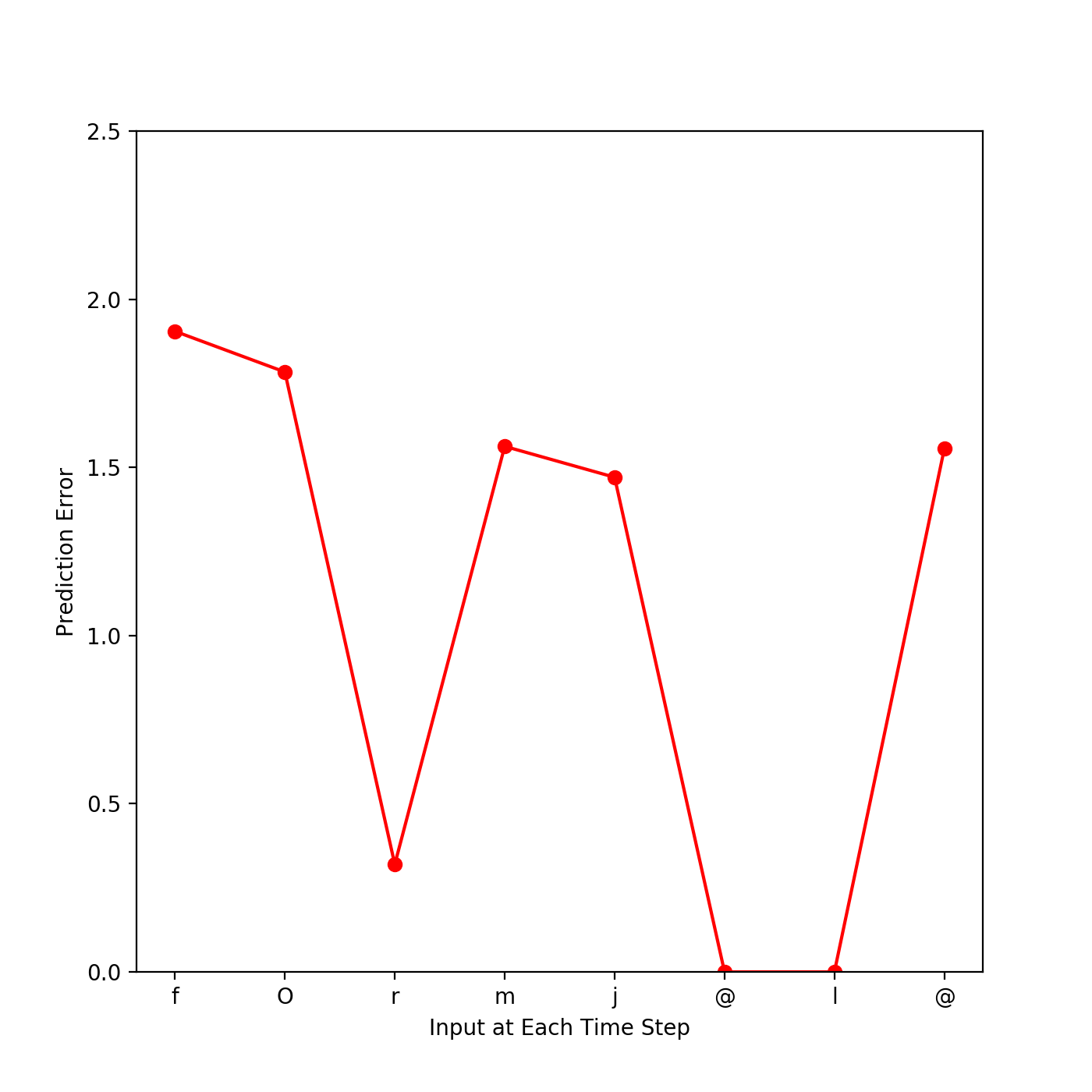
Test plotting
pre_Model.Test_Plot(pronunciation= <str>, file_Suffix=<str>) post_Model.Test_Plot(pronunciation= <str>, file_Suffix=<str>)- Parameters
test_Pronunciation- Determine a phoneme string will be tested.
file_Suffix- Determine the suffix of exported file name
- Parameters
- Result example
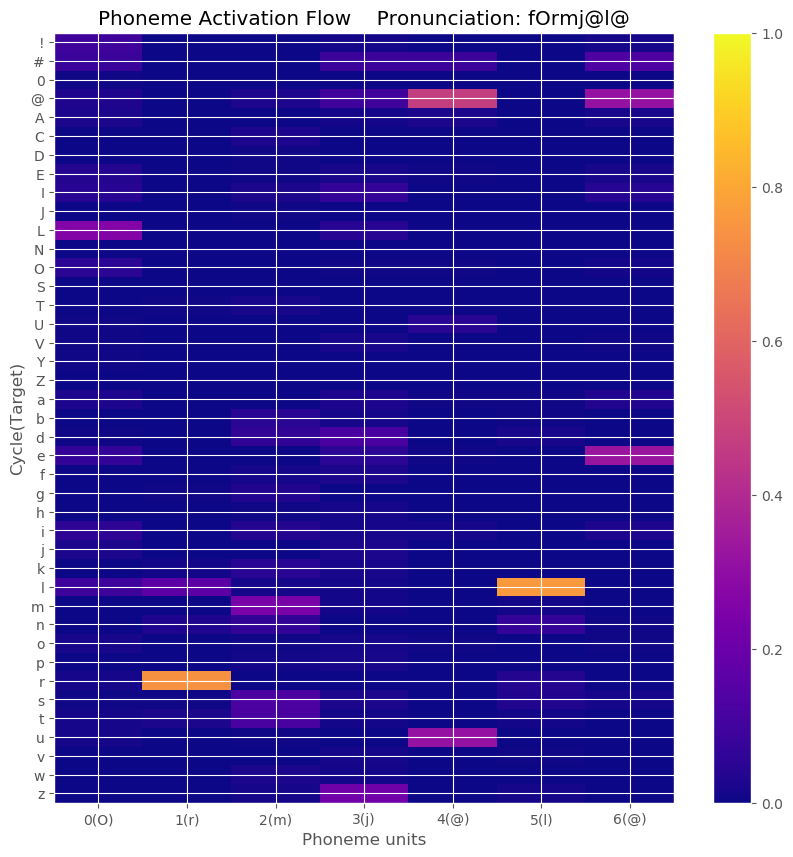
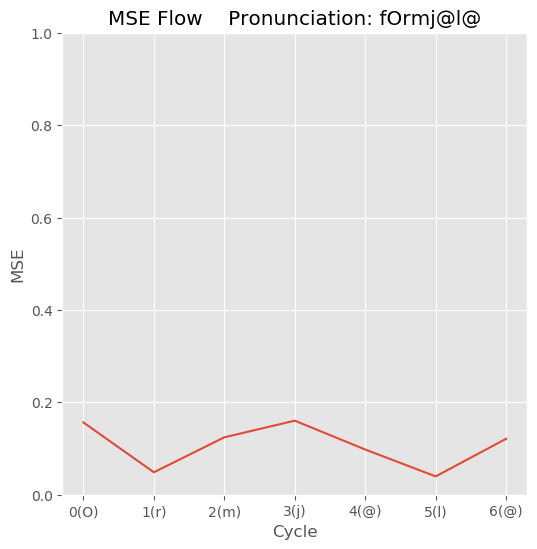
-
-
Phase test
-
Load both of two pre and post addition models.
pre_Model = Model( hidden_Unit= <int>, output_Function= <Sigmoid or Softmax>, lexicon_File= <path>, additional_Lexicon_File= <path>, weight_File= <path>, use_Frequency= <bool> ) post_Novel1_Day1_Model = Model( hidden_Unit= <int>, output_Function= <Sigmoid or Softmax>, lexicon_File= <path>, additional_Lexicon_File= <path>, weight_File= <path>, use_Frequency= <bool> ) post_Novel1_Day2_Model = Model( hidden_Unit= <int>, output_Function= <Sigmoid or Softmax>, lexicon_File= <path>, additional_Lexicon_File= <path>, weight_File= <path>, use_Frequency= <bool> ) post_Novel2_Day1_Model = Model( hidden_Unit= <int>, output_Function= <Sigmoid or Softmax>, lexicon_File= <path>, additional_Lexicon_File= <path>, weight_File= <path>, use_Frequency= <bool> ) post_Novel2_Day2_Model = Model( hidden_Unit= <int>, output_Function= <Sigmoid or Softmax>, lexicon_File= <path>, additional_Lexicon_File= <path>, weight_File= <path>, use_Frequency= <bool> )weight_Fileis located in './Results/<Use_Frequency>/Weight'
-
Type following command
Phase_Test( pre_Model= pre_Model, post_Novel1_Day1_Model= post_Novel2_Day2_Model, post_Novel1_Day2_Model= post_Novel2_Day2_Model, post_Novel2_Day1_Model= post_Novel2_Day2_Model, post_Novel2_Day2_Model= post_Novel2_Day2_Model, tag= <str>, export_Path= <path> )- Result file is
<export_Path>/Result_Data<tag>.txt.
- Result file is
-
Trace simulations were conducted in C (available at http://magnuson.psy.uconn.edu/wp-content/uploads/sites/1140/2015/01/ctrace.zip). Code used for simulations is forthcoming.