3D MRI Brain Tumor Segmentation Using Autoencoder Regularization
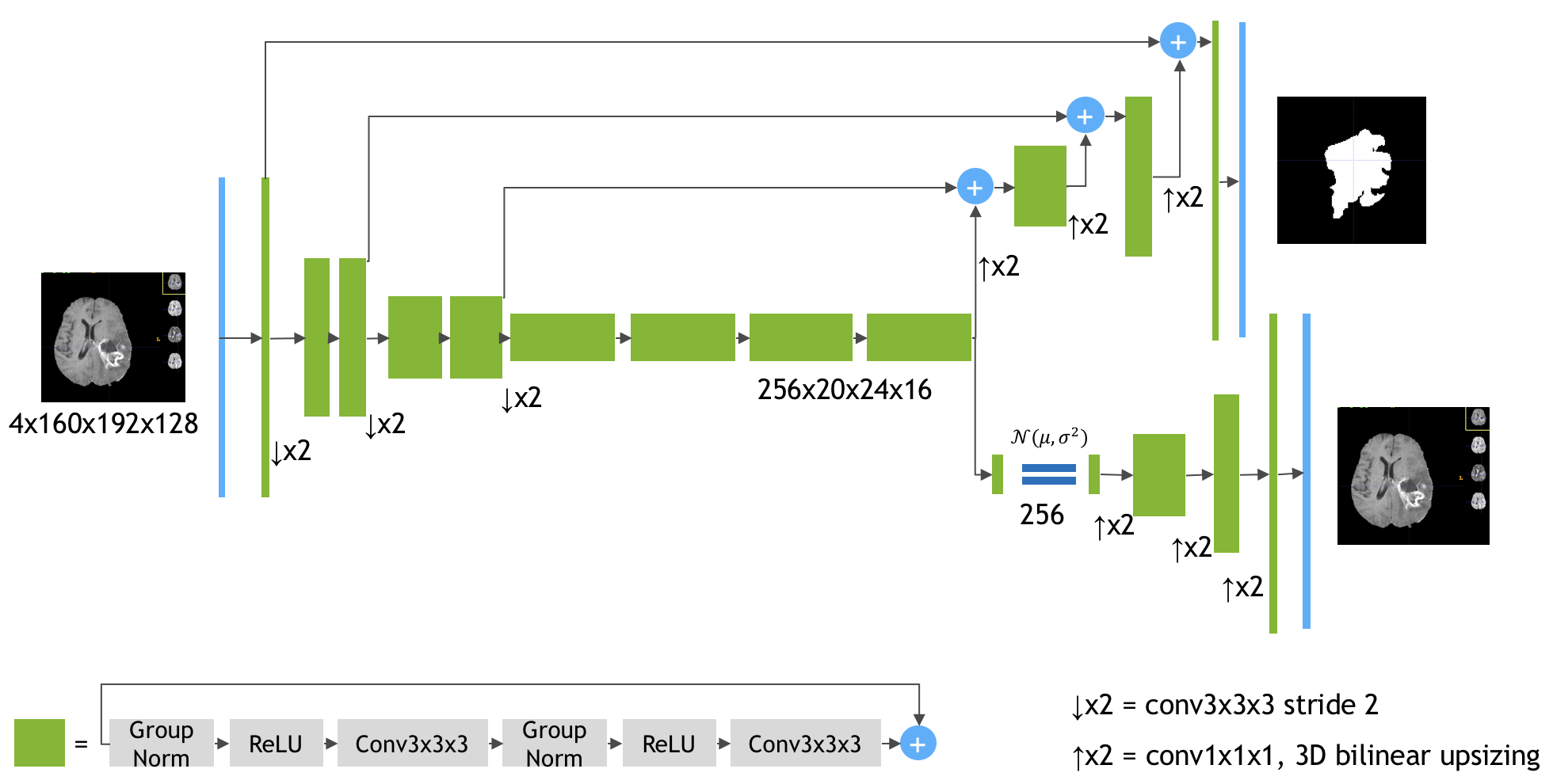
The Model ArchitectureSource: https://arxiv.org/pdf/1810.11654.pdf
Keras implementation of the paper 3D MRI brain tumor segmentation using autoencoder regularization by Myronenko A. (https://arxiv.org/abs/1810.11654). The author (team name: NVDLMED) ranked #1 on the [BRATS 2018](https://www.med.upenn.edu/sbia/brats2018/) leaderboard using the model described in the paper.
This repository contains the model complete with the loss function, all implemented end-to-end in Keras. The usage is described in the next section.
Usage
-
Download the file
model.pyand keep in the same folder as your project notebook/script. -
In your python script, import
build_modelfunction frommodel.py.from model import build_model
It will download an additional script needed for the implementation,
group_norm.py, which contains keras implementation for the group normalization layer. -
Make sure the input MRI scans you are going to feed have channels first format with height, width and depth all divisible by 24, i.e., 16. This is to ensure correct output shape according to the model.
-
Now to create the model, simply run:
model = build_model(input_shape)
where,
input_shapeis a 4-tuple (channels, Height, Width, Depth).
Issues
If you encounter any issue or have a feedback, don't hesitate to raise an issue.