This repository contains benchmarks comparing the performance of different libraries when reading various file formats used in theoretical and computational chemistry and biology. The benchmarks only test opening a file and reading it in full, not running analysis on the resulting trajectory.
Libraries compared:
git clone https://github.com/chemfiles/benchmarks
cd benchmarks
./install-benchmarks.sh
./run-benchmarks.shDifferent libraries are able to read different formats, and can sometimes do more work when opening and reading files.
We benchmark the code running from Python, even if some libraries have other languages interfaces.
We use the version of each software as provided in conda, without special tuning for a given system.
When in doubt, run benchmark using with your own code and use cases!
In most of the tested cases, chemfiles is faster than other softwares. Exceptions to this seems to be the XTC and DCD readers in MDAnalysis, when running on macOS.
Each library is tested with multiple format (text formats, direct reading of compressed text, and binary formats). For each library, the time required to open and read the full trajectory is reported; as well as the timing ratio with respect to chemfiles (higher is slower).
This graph shows the results on Linux:
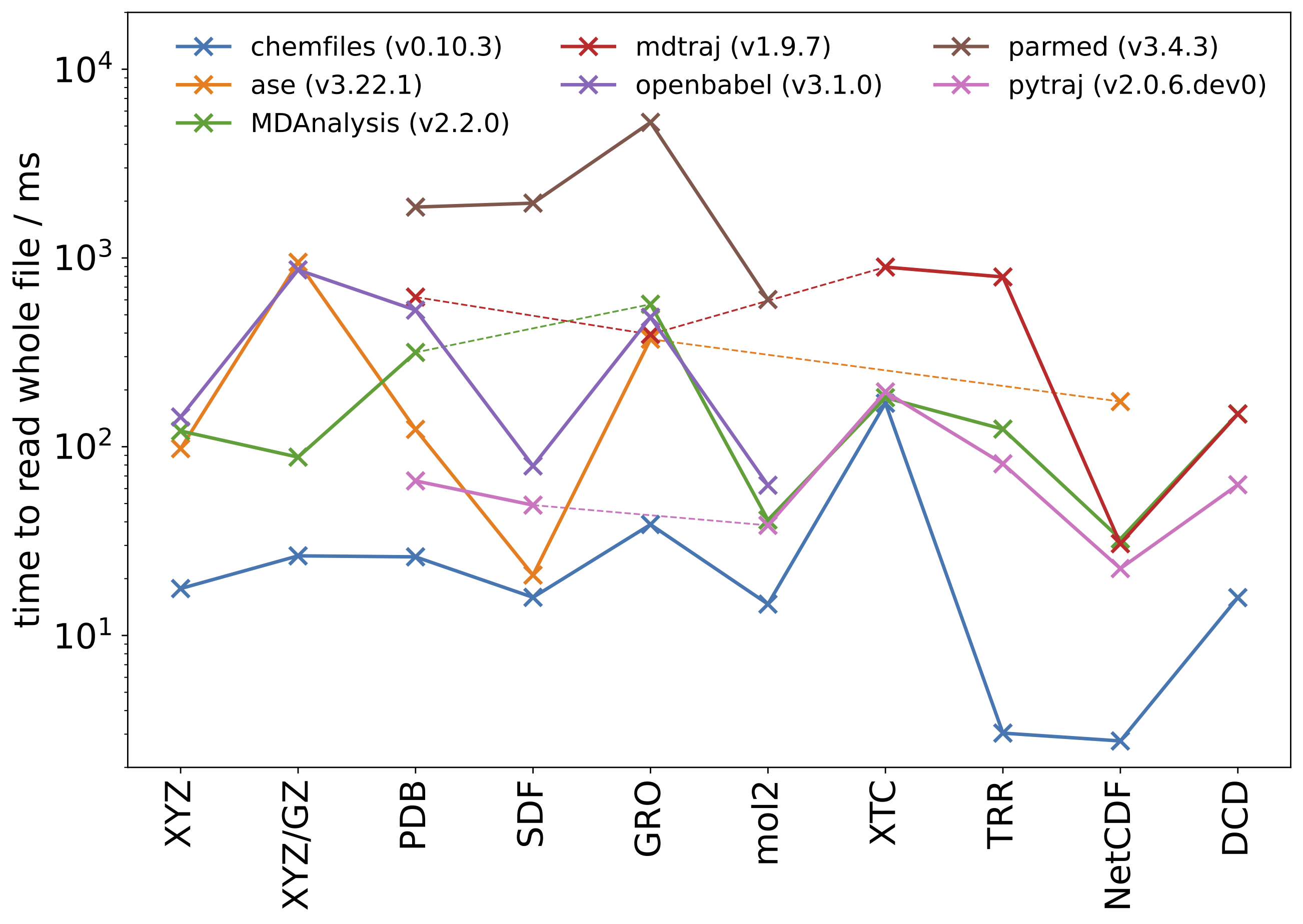
All timings are in milliseconds.
| file | chemfiles (v0.10.3) | ase (v3.22.1) | MDAnalysis (v2.2.0) | mdtraj (v1.9.7) | openbabel (v3.1.0) | parmed (v3.4.3) | pytraj (v2.0.6) |
|---|---|---|---|---|---|---|---|
| water.xyz | 4.87 | 46.00 - 9.5x | 18.52 - 3.8x | 44.57 - 9.2x | |||
| water.xyz.gz | 12.75 | 505.79 - 39.7x | 25.41 - 2.0x | 411.59 - 32.3x | |||
| 1vln-triclinic.pdb | 11.47 | 66.54 - 5.8x | 102.12 - 8.9x | 334.51 - 29.2x | 302.76 - 26.4x | 984.93 - 85.9x | 20.38 - 1.8x |
| lmsd.sdf | 5.19 | 8.62 - 1.7x | 41.62 - 8.0x | 984.03 - 189.4x | 7.04 - 1.4x | ||
| vesicles.gro | 12.91 | 181.34 - 14.0x | 311.04 - 24.1x | 215.69 - 16.7x | 146.50 - 11.4x | 2707.65 - 209.7x | |
| molecules.mol2 | 5.18 | 15.72 - 3.0x | 29.02 - 5.6x | 325.65 - 62.8x | 11.08 - 2.1x | ||
| ubiquitin.xtc | 79.41 | 58.10 - 0.73x | 473.29 - 6.0x | 69.61 - 0.88x | |||
| ubiquitin.trr | 1.19 | 24.43 - 20.5x | 432.94 - 363x | 18.85 - 15.8x | |||
| water.nc | 0.92 | 36.44 - 39.4x | 2.02 - 2.2x | 4.21 - 4.55x | 7.16 - 7.8x | ||
| adk.dcd | 7.18 | 5.39 - 0.75x | 85.56 - 11.9x | 12.74 - 1.8x |
All timings are in milliseconds.
| file | chemfiles (v0.10.3) | ase (v3.22.1) | MDAnalysis (v2.2.0) | mdtraj (v1.9.7) | openbabel (v3.1.0) | parmed (v3.4.3) | pytraj (v2.0.6) |
|---|---|---|---|---|---|---|---|
| water.xyz | 17.72 | 97.73 - 5.5x | 121.17 - 6.8x | 143.75 - 8.1x | |||
| water.xyz.gz | 26.41 | 948.20 - 35.9x | 88.07 - 3.3x | 865.02 - 32.8x | |||
| 1vln-triclinic.pdb | 26.09 | 123.60 - 4.7x | 315.89 - 12.1x | 620.56 - 23.8x | 529.36 - 20.3x | 1860.58 - 71.3x | 65.92 - 2.5x |
| lmsd.sdf | 15.92 | 20.88 - 1.3x | 79.10 - 5.0x | 1952.52 - 122.7x | 49.05 - 3.1x | ||
| vesicles.gro | 38.72 | 371.38 - 9.6x | 568.72 - 14.7x | 393.73 - 10.2x | 485.10 - 12.5x | 5229.44 - 135.1x | |
| molecules.mol2 | 14.66 | 40.90 - 2.8x | 62.50 - 4.3x | 602.32 - 41.1x | 38.32 - 2.6x | ||
| ubiquitin.xtc | 169.95 | 181.97 - 1.0x | 895.10 - 5.3x | 195.14 - 1.2x | |||
| ubiquitin.trr | 3.04 | 123.98 - 40.8x | 793.42 - 261.0x | 81.19 - 26.7x | |||
| water.nc | 2.76 | 173.51 - 62.8x | 32.44 - 11.8x | 30.65 - 11.1x | 22.63 - 8.2x | ||
| adk.dcd | 15.87 | 149.44 - 9.4x | 149.02 - 9.4x | 62.90 - 4.0x |
Improvements to the benchmarks and more files to be tested are very welcome!