IPython Notebook for downloading and analyzing data from the manuscript: "Indication of family-specific DNA methylation patterns in developing oysters"
The repository includes a IPython notebook (.ipynb file) that can be downloaded and interactively executed. The code in the IPython notebook will download raw data and process data such that figures in the manuscript are reproduced (in theory).
To execute the IPython Notebook in its entirety you will need:
- IPython - install instructions
- BSMAP - install instructions
- bedtools - install instructions
- R - install instructions
- rpy2 (interface to R from Python) - install instructions
Sofware versions originally used in this analyses (on Mac OS X v10.7.5) are as follows:
- IPython: 2.3.0
- BSMAP: 2.74
- bedtools: 2.17.0
- R: 3.1.1
- rpy2: 2.5.0
Note the current version of the IPython Notebook can be viewed (not interactive) in any web browser at: http://nbviewer.ipython.org/github/che625/olson-ms-nb/blob/master/BiGo_dev.ipynb
##Instructions
1) Download the repository zip file to a local directory and uncompress. This can be done by clicking on the link in the right sidebar or directly downloading: https://github.com/che625/olson-ms-nb/archive/master.zip
2) Launch IPython from the repository primary directory. For example, using Terminal on MacOSX.
$ cd /Desktop/olson-ms-nb
$ ipython notebook
This will launch IPython in your web browser.
3) Open notebook by clicking on BiGo_dev.ipynb. This will open a new tab in your browser.
4) Execute cells in notebook The first section of the notebook includes code to download raw data in wd subdirectory. In theory, assuming all dependencies are installed
- BSMAP
- bedtools
- R
- rpy2 (interface to R from Python)
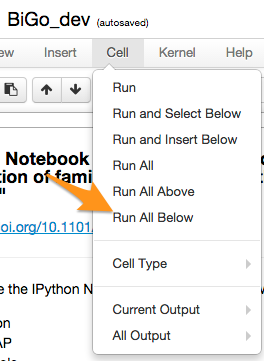
you could edit the cell near the top to provide the location of BSMAP on your machine, then run all cells (see screenshot). Raw data will be downloaded, and analyses carried out, producing figures (2) in manuscript. Please note data is very large (>20 GB) and analyses will take several hours depending on your machine.
In practice, you can execute cells individually with shift-enter.
We are actively trying to improve this realizing that we are likely missing dependancies, etc. Any suggestions and feedback is welcome.