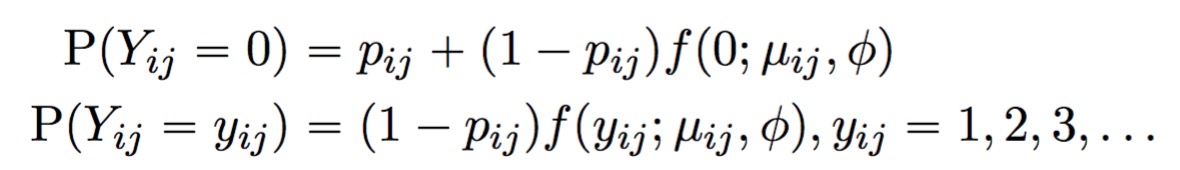twosigma is an R package for differential expression (DE) analysis and gene set testing (GST) in single-cell RNA-seq (scRNA-seq) data. At the gene-level, DE can be assessed by fitting our proposed TWO-component SInGle cell Model-based Association method (TWO-SIGMA). The first component models the drop-out probability with a mixed effects logistic regression, and the second component models the (conditional) mean read count with a mixed-effects log-linear negative binomial regression. Our approach thus allows for both excess zero counts and overdispersed counts while also accommodating dependency in both drop-out probability and mean mRNA abundance. TWO-SIGMA is especially useful in its flexibility to analyze DE beyond a two-group comparison while simultaneously controlling for additional subject-level or cell-level covariates including batch effects. At the set level, the package can perform competitive gene set testing using our proposed TWO-SIGMA-G method.
We recommend installing from Github for the latest version of the code:
install.packages("devtools")
devtools::install_github("edvanburen/twosigma")
library(twosigma)TWO-SIGMA is based on the following parameterization of the negative binomial distribution:
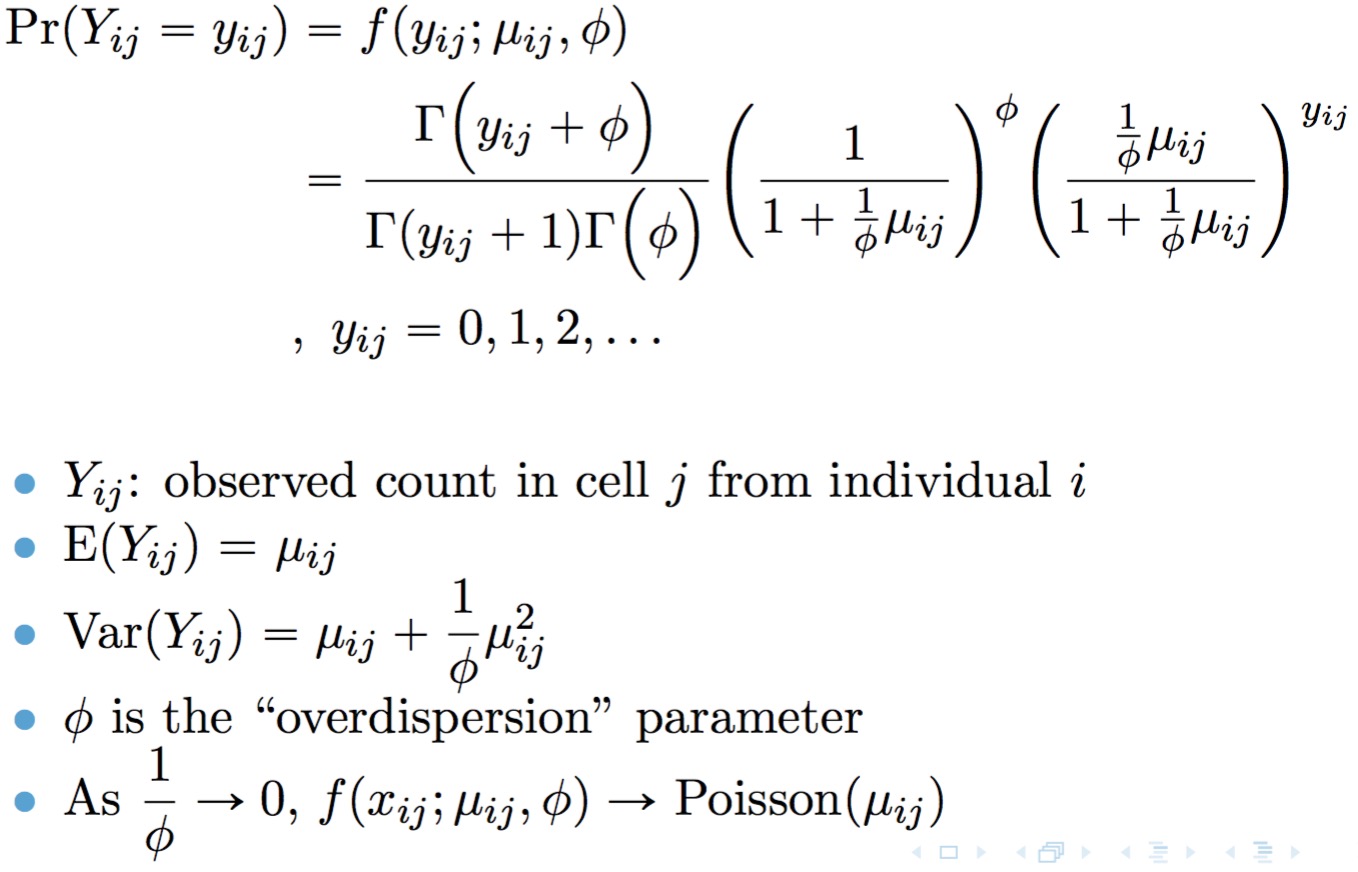
A point mass at zero is added to the distribution to account for dropout. The result is the probability mass function for the zero-inflated negative binomial distribution:
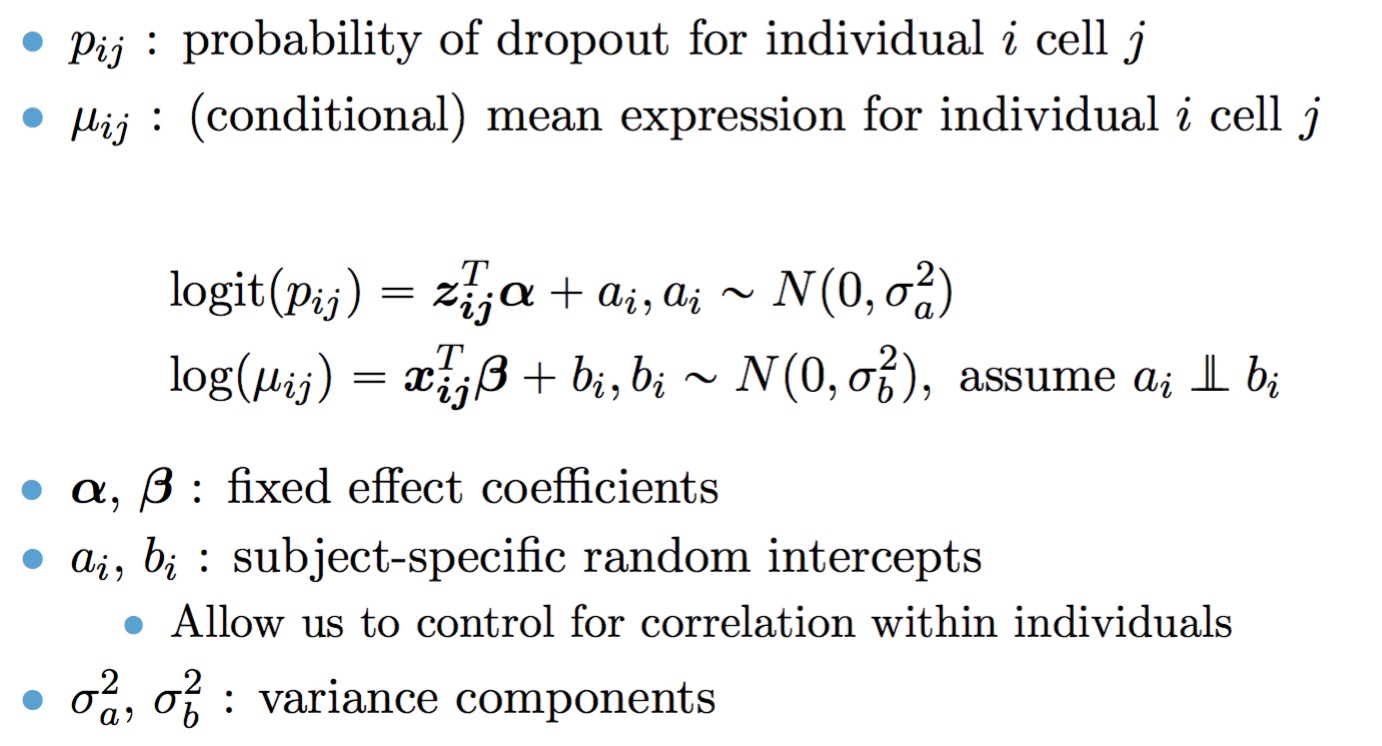
The full TWO-SIGMA specification is therefore as follows:
The workhorse function is twosigma, which can be easiest called as
twosigma(count, mean_covar, zi_covar,id)- count: A vector of non-negative integer counts. No normalization is done.
- mean_covar: A matrix (such as from model.matrix) of covariates for the (conditional) mean model without an intercept term. Columns give covariates and the number of rows should correspond to the number of cells.
- zi_covar: A matrix (such as from model.matrix) of covariates for the zero-inflation model without an intercept term. Columns give covariates and the number of rows should correspond to the number of cells.
- id: Vector of individual-level ID's (length equal to the total number of cells). Used for random effect prediction and the ad hoc method and is currently required even if neither is being used.
By default, we employ our ad hoc procedure to determine if random effects are needed. If users wish to specify their own random effect specifications, they can set adhoc=FALSE, and use the following inputs:
- mean_re: Should random intercept terms be included in the (conditional) mean model?
- zi_re: Should random intercept terms be included in the zero-inflation model?
If adhoc=TRUE, mean_re and zi_re are ignored and a warning is printed.
If users wish to customize the random effect specification, they may do so via the function twosigma_custom , which has the following basic syntax:
twosigma_custom(count, mean_form, zi_form, id)- count: A vector of non-negative integer counts. No normalization is done. Batch can be controlled for by inclusion in the design matrices.
- mean_form a two-sided formula for the (conditional) mean model. Left side specifies the response and right side includes fixed and random effect terms. Users should ensure that the response has the name "count", e.g.
mean_form = count ~ 1 - zi_form a one-sided formula for the zero-inflation model including fixed and random effect terms, e.g.
~ 1 - id: Vector of individual-level ID's. Used for random effect prediction.
Some care must be taken, however, because these formulas are used directly. It is therefore the user's responsibility to ensure that formulas being inputted will operate as expected. Syntax is identical to the lme4 package.
For example, each of the following function calls reproduces the default TWO-SIGMA specification with random intercepts in both components:
twosigma(count=counts, mean_covar=mean_covar_matrix, zi_covar=zi_covar_matrix, mean_re = TRUE, zi_re = TRUE, id=id,adhoc=F)
twosigma_custom(count=counts, mean_form=count~mean_covar_matrix+(1|id),zi_form=~zi_covar_matrix+(1|id),id=id)If users wish to jointly test a fixed effect using the twosigma model with a non-custom specification via the likelihood ratio test, they may do so using the lr.twosigma or lr.twosigma_custom functions:
lr.twosigma(count, mean_covar, zi_covar, contrast, mean_re = TRUE,zi_re = TRUE, disp_covar = NULL,adhoc=TRUE)
lr.twosigma_custom(count, mean_form_alt, zi_form_alt, mean_form_null, zi_form_null, id, lr.df)- contrast: Either a string indicating the column name of the covariate to test or an integer referring to its column position in BOTH the mean_covar and zi_covar matrices. If an integer is specified there is no check that it corresponds to the same covariate in both the mean_covar and zi_covar matrices.
- lr.df If custom formulas are input users must provide the degrees of freedom from which the likelihood ratio p-value can be calculated. Must be a non-negative integer.
The lr.twosigma function assumes that the variable being tested is in both components of the model (and thus that the zero-inflation component exists and contains more than an Intercept). Users wishing to do fixed effect testing in other cases can use the lr.twosigma_custom function with custom formulas or construct the test themselves using two calls to twosigma or twosigma_custom. The formula inputs mean_form_alt , mean_form_null, zi_form_alt, and zi_form_null should be specified as in the lr.twosigma_custom function and once again users must ensure custom formulas represent a valid likelihood ratio test.
As mentioned in the paper, we mention a method that can be useful in selecting genes that may benefit from the inclusion of random effect terms. This method fits a zero-inflated negative binomial model without random effects and uses a one-way ANOVA regressing the Pearson residuals on the individual ID to look for differences between individuals.
adhoc.twosigma(count, mean_covar, zi_covar, id)The p-value from the ANOVA F test is returned, and can be used as a screening for genes that are most in need of random effects. This functionality is built into the twosigma function so users likely do not need to call directly themselves.
Analogously, there are two workhorse function for gene set testing: twosigmag and twosigmag_custom.
The adhoc procedure is not recommended for use in gene set testing. This is because geneset testing relies on a common gene-level null hypothesis being tested. When some genes have random effects and others do not, it is not clear that this requirement is met. Arguments which require more explanation over above are given as follows:
- count_matrix: a matrix of counts, where rows are genes and columns are cells
- index_test: Either a numeric vector, or a list of numeric vectors, corresponding to indices (row numbers) belonging to the test set(s) of interest.
- index_ref: An optional numeric vector, or list of numeric vectors, corresponding to indices (row numbers) belonging to desired reference set(s). Most users should not need to modify this option.
- all_as_ref: Should all genes not in the test set be taken as the reference set? Defaults to true. If FALSE, a random sample of size identical to the size of the test set is taken as the reference.
- allow_neg_corr: Should negative correlations be allowed for a test set? By default negative correlations are set to zero to be conservative. Most users should not need to modify this option.
#-----------------Simulate data-----------------#
#Set parameters for the simulation
nind<-10
ncellsper<-rep(100,nind)
sigma.a<-.1
sigma.b<-.1
alpha<-c(1,0,-.5,-2)
beta<-c(2,0,-.1,.6)
phi<-.1
id.levels<-1:nind
# Simulate some individual level covariates as well as cell-level Cellular Detection Rate (CDR)
t2d_ind<-rbinom(nind,1,p=.4)
t2d_sim<-rep(t2d_ind,times=ncellsper)
nind<-length(id.levels)
id<-rep(id.levels,times=ncellsper)
cdr_sim<-rbeta(sum(ncellsper),3,6) #Is this good distn
age_sim_ind<-sample(c(20:60),size=nind,replace = TRUE)
age_sim<-rep(age_sim_ind,times=ncellsper)
#Construct design matrices
Z<-cbind(scale(t2d_sim),scale(age_sim),scale(cdr_sim))
colnames(Z)<-c("t2d_sim","age_sim","cdr_sim")
X<-cbind(scale(t2d_sim),scale(age_sim),scale(cdr_sim))
colnames(X)<-c("t2d_sim","age_sim","cdr_sim")
sim_dat<-simulate_zero_inflated_nb_random_effect_data(ncellsper,X,Z,alpha,beta,phi,sigma.a,sigma.b,
id.levels=NULL)
#-----------------------------------------------#
#--- Fit TWO-SIGMA to simulated data
id<-sim_dat$id
counts<-sim_dat$Y
fit<-twosigma(count=counts,zi_covar=Z,mean_covar = X,id=id,mean_re=TRUE,zi_re=TRUE,adhoc=F)
fit2<-twosigma_custom(count=counts, mean_form=count~X+(1|id),zi_form=~Z+(1|id),id=id)
#fit and fit2 are the same
summary(fit)
summary(fit2)
confint(fit)
#--- Fit TWO-SIGMA without a zero-inflation component
fit_noZI<-twosigma(count=counts,zi_covar=0,mean_covar = X,id=id,mean_re=F,zi_re=F,adhoc=F)
fit_noZ2I<-twosigma_custom(count=counts,zi_form=~0,mean_form=count~X,id=id)
#--- Fit TWO-SIGMA with an intercept only zero-inflation component and no random effects
fit_meanZI<-twosigma(count=counts,zi_covar=1,mean_covar = X,id=id,mean_re=F,zi_re=F,adhoc=F)
fit_meanZI2<-twosigma_custom(count=counts, mean_form=count~X+(1|id),zi_form=~1,id=id)
summary(fit_noZI)
summary(fit_meanZI)
# Perform Likelihood Ratio Test on variable "t2d_sim"
lr.fit<-lr.twosigma(contrast="t2d_sim",count=counts,mean_covar = X,zi_covar=Z,id=id)
lr.fit$p.val
summary(lr.fit$fit_alt)
lr.fit_custom<-lr.twosigma_custom(count=counts,mean_form_alt=count~X, zi_form_alt=~Z, mean_form_null=count~X[,-1],zi_form_null=~Z[,-1],id=id,lr.df=2)
lr.fit_custom$p.val
summary(lr.fit_custom$fit_alt)
# Perform adhoc method to see if random effects are needed
adhoc.twosigma(count=counts,zi_covar=Z,mean_covar=X,id=id)
#--------------------------------------------------
# Perform Gene-Set Testing
#--------------------------------------------------
#----------
# First, simulate some DE genes
sim_dat2<-matrix(nrow=10,ncol=sum(ncellsper))
beta<-c(2,0,-.1,.6)
beta2<-c(2,.2,-.1,.6)
for(i in 1:nrow(sim_dat2)){
if(i<5){
sim_dat2[i,]<-simulate_zero_inflated_nb_random_effect_data(ncellsper,X,Z,alpha,beta2,phi,sigma.a,sigma.b=.5,
id.levels=NULL)$Y
}else{
sim_dat2[i,]<-simulate_zero_inflated_nb_random_effect_data(ncellsper,X,Z,alpha,beta,phi,sigma.a,sigma.b,
id.levels=NULL)$Y
}
}
gst<-twosigmag(sim_dat2,index_test = 6:10,index_ref = 1:5,contrast="t2d_sim",mean_covar=X,zi_covar=Z
,id=id,mean_re=FALSE,zi_re=FALSE)
names(gst)
gst$set_p.val
gst<-twosigmag_custom(sim_dat2,index_test = 6:10,index_ref = 1:5,mean_form_alt =count~t2d_sim+cdr_sim+age_sim
,zi_form_alt = ~t2d_sim+cdr_sim+age_sim,mean_form_null = count~cdr_sim+age_sim,zi_form_null = ~cdr_sim+age_sim,id=id,lr.df=2)
names(gst)
gst$set_p.val