- Please download and install Bandage (https://rrwick.github.io/Bandage/) on your local computer
- The rest of the software will be available in your cloud session:
odgito subset and manipulate genome graphsvgto map reads, count read depth, and call variantsbcftoolsto work with VCF files, call variantssamtoolsto work with BAM files
Some steps take a long time that is prohibitive during a workshop. Therefore,
long running commands are written to use a small subsample of data that allows
you to run the command very fast and understand the tool parameters and output.
Later steps then use prepared intermediate files that reside in /data/ to
provide the full results.
You may download the data at https://doi.org/10.5281/zenodo.10794014.
The Human Pangenome Reference Consortium released the first draft human pangenome reference built from 94 genetically diverse assemblies. This reference uses a graph data structure to represent the known genetic variation at any given locus. Every HPRC genome is a path through the graph and additional genomes are represented as paths that pass through a combination of HPRC genomes.
Genome graphs can be built in various way. The HPRC graph was built from a
multiple genome alignment involving 94 assemblies, but graphs can also be built
from VCF files. Run vg autoindex to see some options for building graph
genomes.
We will open the HPRC graph and inspect some subregions of chromosome 22 with a
tool called Bandage. First, we already extracted chromosome 22 from the whole
genome graph in the file called hprc-chrom22.odgi. We can use odgi
(https://github.com/pangenome/odgi) to ask for specific subgraphs of the human
pangenome. For example, this extracts the subgraph of the DGCR6 gene:
Command 1
odgi extract -d 0 -i /data/hprc-chrom22.odgi -c 10 -E -r GRCh38.chr22:18906028-18912088 -o - | odgi view -g -i - > DGCR6.gfaKey parameters:
-iinput graph-rregion interval along GRCh38.chr22 path to extract (path:start-end)-Eextract the subgraph to include nodes and paths from other genomes that touch on GRCh38.chr22. Without this argument, the results will be a simple linear graph without variants-cnumber of surrounding nodes to extract around the specified region. This gives the extracted subgraph a bit more context
Download the file DGCR6.gfa to your own computer by running this on your local UNIX computer.
You may ignore -i your_key if you have a username and password:
Command 2
scp -i your_key your_username@52.69.106.213:DGCR6.gfa .Windows users will have to install PuTTY and use pscp:
Command 3
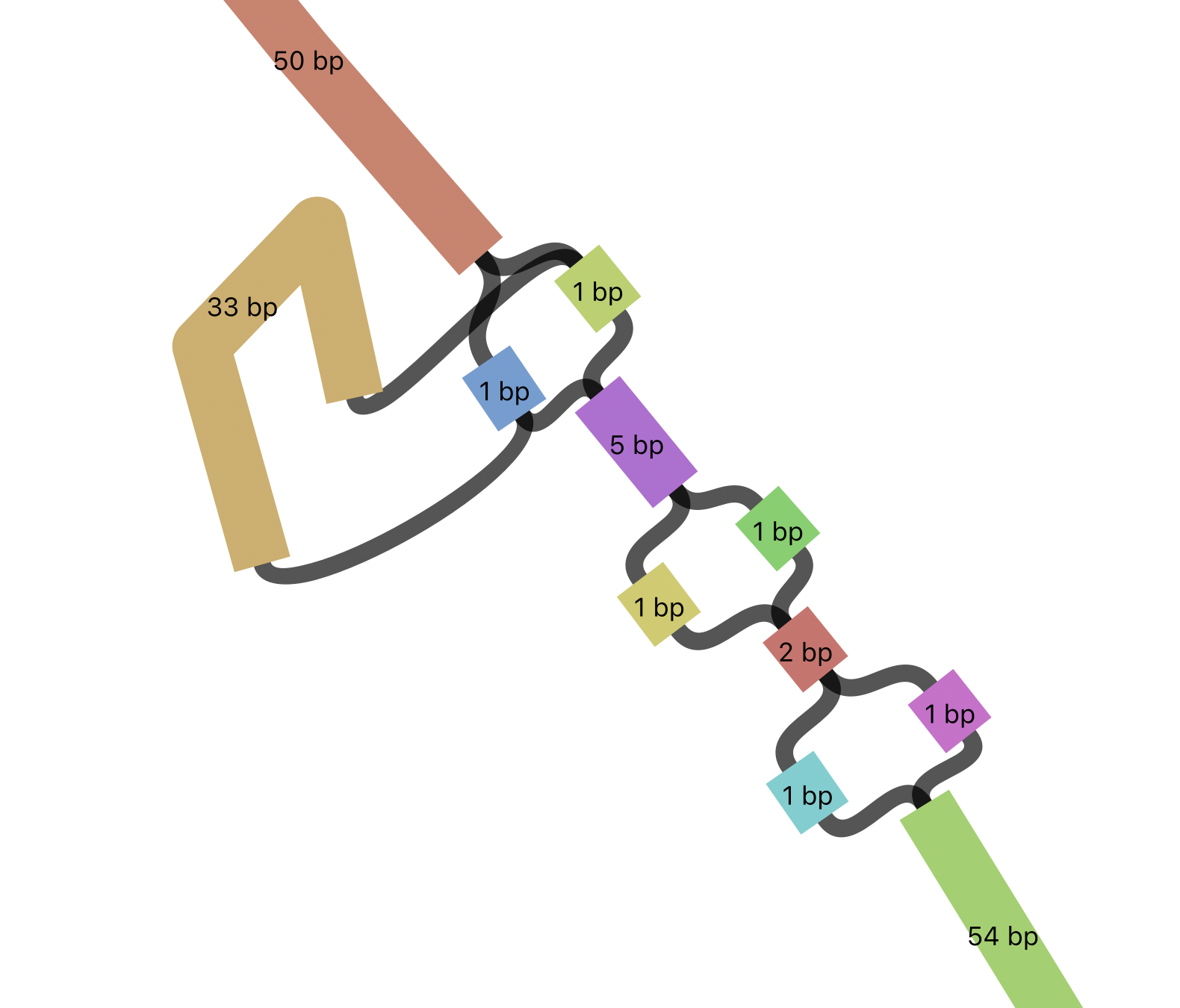
pscp -scp your_username@52.69.106.213:DGCR6.gfa .Once you have the file, load it in Bandage (File -> Load graph, Draw graph). You should see a long, mostly linear structure. However, you should also see small bubbles along this gene, representing SNVs and indels. In Bandage, use the find node field in the top right of the Bandage window to search for node 270054. You should see a polymorphic region containing a 33 bp indel, and 3 consecutive SNVs within a very short region.
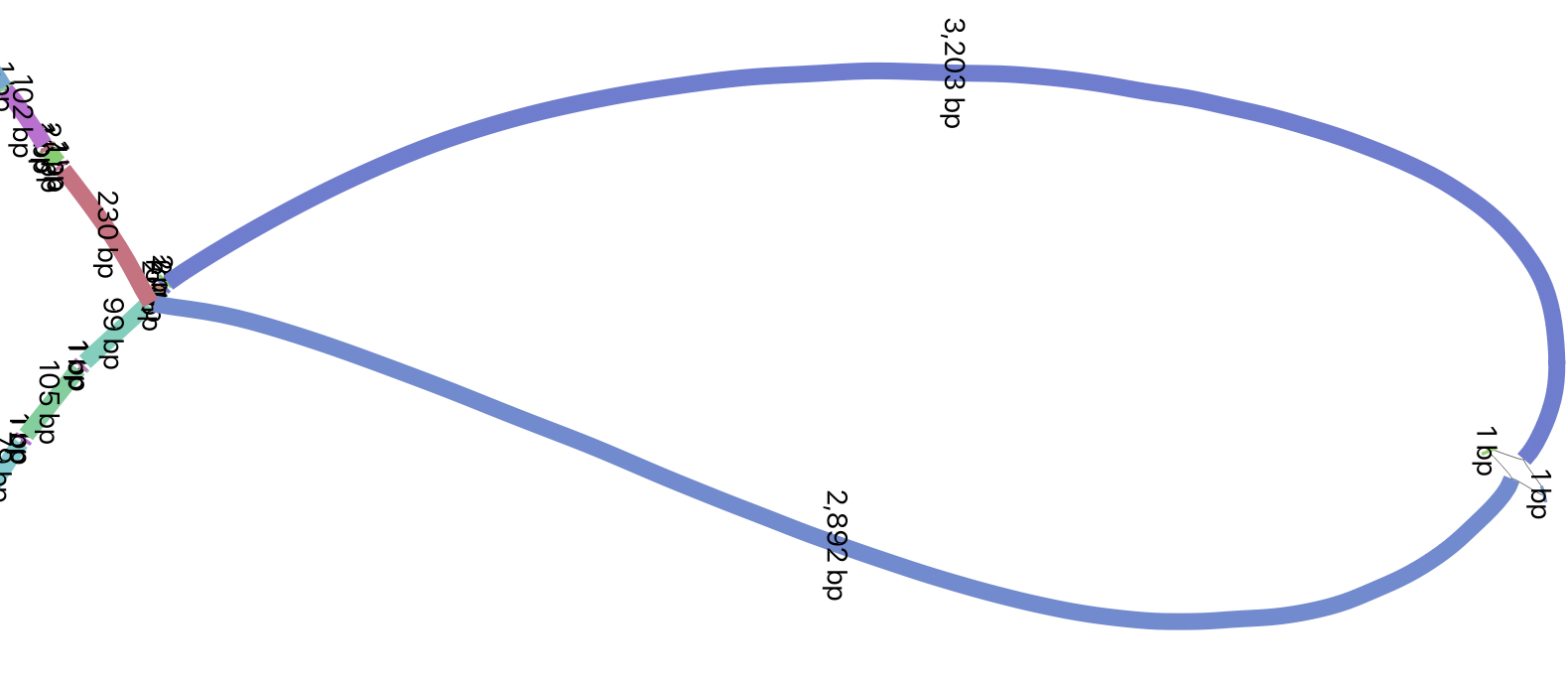
Let’s find a larger structural variant, in this case a 6 kbp LINE1 insertion:
Command 4
odgi extract -d 0 -i /data/hprc-chrom22.odgi -c 100 -E -r GRCh38.chr22:26101249-26121249 -o - | odgi view -g -i - > L1.gfaAgain, download and open the file L1.gfa (just like before, but replace
DGCR6.gfa with L1.gfa) in Bandage and find node 438987. You should see
something similar to this:
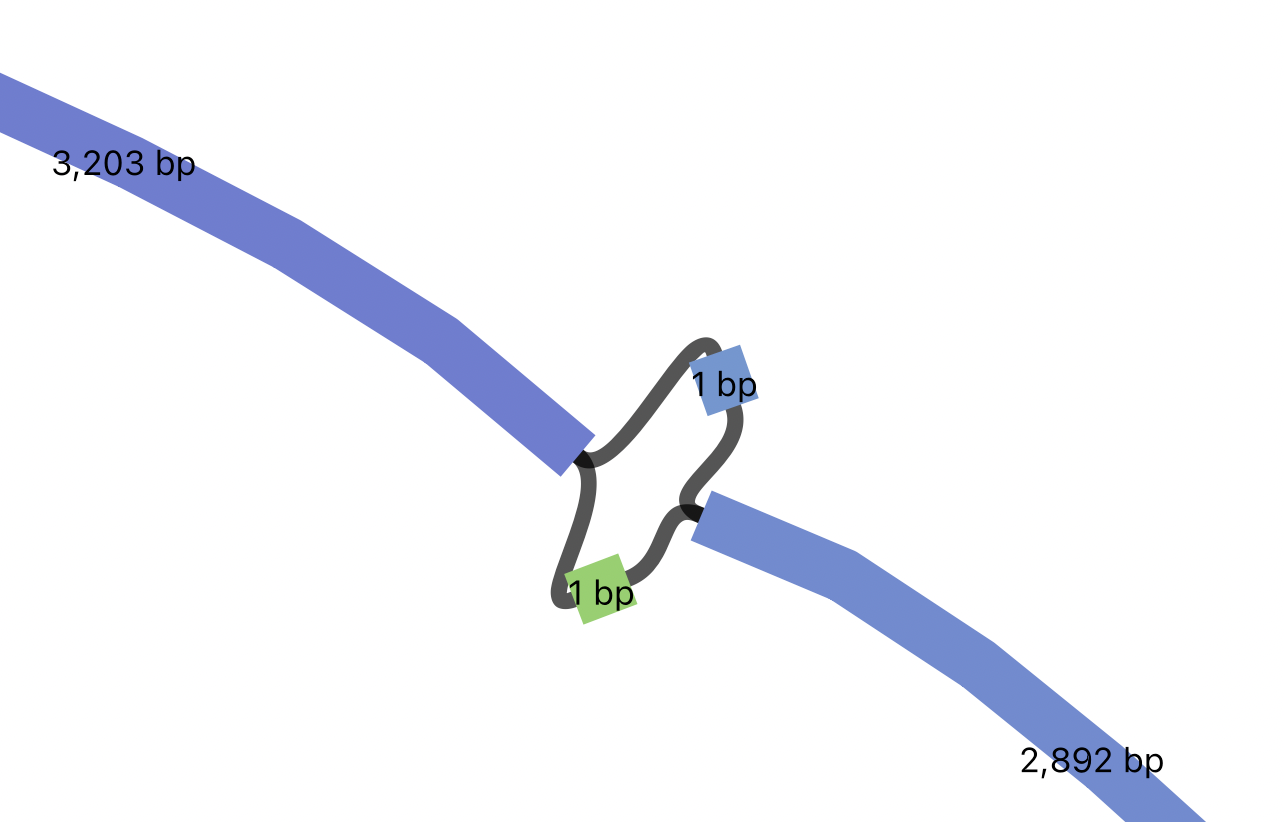
If you zoom in, you should see a small bubble on the L1, representing a SNV nested within the L1 insertion. Note how the graph represents the nested SNP within the LINE1 insertion as a bubble within a bubble:
We may use the human pangenome reference similarly to how we use GRCh38, to
align existing short read data. Let’s align a whole genome sequencing dataset of
NA12878 to chr22. We can do this using the graph aligner vg giraffe
(https://github.com/vgteam/vg), and the appropriate index files which can be
downloaded at https://github.com/human-pangenomics/hpp_pangenome_resources.
Here, we align a small sample of reads from chromosome 22 to a graph.
Command 5
vg giraffe -p -i -f /data/NA12878.chr22.sample.fq.gz -Z /data/hprc-chrom22.giraffe.gbz -m /data/hprc-chrom22.min -d /data/hprc-chrom22.dist -o gam > NA12878.chr22.sample.gamKey parameters:
-pshow progress-iinput reads are an interleaved paired-end FASTQ file-finput FASTQ file-Zinputgiraffegraph (created withvg autoindex)-mminimizer index, a table that lists where the position of k-mers are in the graph-ddistance index, a table that letsgiraffequickly calculate the minimum distance inbpbetween any two nodes in the graph-o gafdata format for alignments (can be GAM, GAF or others)
At the end of this operation you should see a message similar to this:
Mapped 10044682 reads across 40 threads in 72.2703 seconds with 1.02009 additional single-threaded seconds. Achieved 1469.49 reads per CPU-second (including output)
As you can see, vg giraffe is very fast!
The GAM is a binary format is analogous to the BAM format. It is space efficient
but not human readable. GAF is a text file format that is more similar to the
SAM file format. While various tools prefer GAF or GAM, vg giraffe allows
users to choosing using the -o gaf/gam command line parameter. You can also
convert between GAM and GAF using vg convert. You can view a record of the GAF
format using:
Command 6
head -n1 /data/NA12878.chr22.sample.gafThere are many similarities with the BAM/SAM format. For example, there are
fields for the read name and its mate, and a mapping quality field. The path
field is analogous to the chromosome field but is more complex. As you can see,
it is an enumeration of nodes instead of a chromosome and the alignments
describe the path in the pangenome on which the read lies. More on the GAF
format:
https://github.com/lh3/gfatools/blob/master/doc/rGFA.md#the-graph-alignment-format-gaf
The aligned whole genome sequencing reads can now be used to genotype variation that is present in the pangenome reference. This approach is particularly useful for genotyping structural variation, which is not accessible to most short read variant callers. To genotype alleles that are present in the graph, we first need to know how many reads align to each allele. We can count this quickly with the following command:
Command 7
vg pack -x /data/hprc-chrom22.giraffe.gbz -g /data/NA12878.chr22.sample.gam -o NA12878.chr22.packThis creates a pack table that describes how many reads align to each node in the pangenome.
Key parameters:
-xinputgiraffegraph-ginput graph alignments fromgiraffe-ooutput node read depth table
We are now ready to call variants using vg call:
Command 8
vg call -r /data/hprc-chrom22.snarls -s NA12878 -k /data/NA12878.chr22.pack /data/hprc-chrom22.giraffe.gbz > NA12878.chr22.hprc.vcfThe output is a VCF file that lists the alleles where NA12878 differs from the GRCh38 path/reference.
Key parameters
-rindex that letsvgquickly find the bubbles in the graph-ssample name to be used in VCF-kinput read depth table from previous step- input
giraffegraph
If you wish to output the genotype of all the alleles, including those that are the same as GRCh38, pass the -a parameter
to vg call. This is useful when working with many samples.
Then, we can quickly summarize the findings with:
Command 9
bcftools stats /data/NA12878.chr22.hprc.vcf | awk -v FS='\t' '$1 == "SN" {print $3,$4}'The output should reproduce the data in this table:
| number of records | 37137 |
| number of SNPs | 26511 |
| number of MNPs | 367 |
| number of indels | 10158 |
| number of others | 263 |
| number of multiallelic sites | 792 |
| number of multiallelic SNP sites | 11 |
|---|
Overall, there are 28,636 insertions and 142,417 SNVs in the graph, of which
10,158 indels and 26,511 SNVs are in NA12878. However, NA12878 likely contains
many rare SNVs and indels that are not present in the pangenome reference and
thus were not called. To recover these, we may still leverage the pangenome
reference to remove reference bias and improve our precision and sensitivity. We
do so by using these HPRC-corrected alignments with traditional callers such as
bcftools, which work on reads aligned to the linear GRCh38 reference.
Since GRCh38 is part of the pangenome, it’s relationship to the HPRC genomes is
described by the graph. Therefore, any alignment that lies on a bubble can be
rewritten as an alignment relative to GRCh38, where the bubble is expressed as
an edit. For example, an alignment that matches 23 bp on GRCh38, 64 bp on HG002
and 44 bp on GRCh38 will be projected to GRCh38 as 23M64I44M. Note that there
is nothing special about GRCh38, we may project onto any genome in the graph
(specified with -p). Project with the following command:
Command 10
vg surject -p GRCh38.chr22 -t 2 -b -x /data/hprc-chrom22.giraffe.gbz /data/NA12878.chr22.sample.gam | samtools sort > NA12878.chr22.hprc.sample.bamThe output file NA12878.chr22.hprc.bam is now a regular BAM file that contains
alignments projected onto GRCh38.
Key parameters:
-ppath onto which to project the alignments. Can be any genome that is part of the graph-tnumber of threads to use, more is faster-boutput BAM-xinputgiraffegraph- input GAM file to surject to BAM
If you check a record in the BAM file, you will see the path named GRCh38.chr22 in the chromosome field:
Command 11
samtools view NA12878.chr22.hprc.sample.bam | head -n1This file can be accepted by tools such as DeepVariant or bcftools. Let’s
call SNVs and indels with bcftools on HPRC-corrected alignments:
Command 12
bcftools mpileup -Ou -f /data/GRCh38.chr22.fa /data/NA12878.chr22.hprc.sample.bam | bcftools call -mv -Ov -o NA12878.chr22.bcftools.vcfLet’s summarize the new results just like before:
Command 13
bcftools stats /data/NA12878.chr22.bcftools.vcf | awk -v FS='\t' '$1 == "SN" {print $3,$4}'Now compare with variants called with bwa mem:
Command 14
bcftools stats /data/NA12878.chr22.bwamem.vcf | awk -v FS='\t' '$1 == "SN" {print $3,$4}'Indeed we find more SNVs and indels that are unique to NA12878 when we use HPRC-corrected alignments.
| Variants | HPRC-corrected | bwa mem | Gain |
|---|---|---|---|
| number of SNPs | 65447 | 67146 | -1699 |
| number of indels | 14172 | 10672 | 3500 |
| number of multiallelic sites | 415 | 349 | 66 |
| number of multiallelic SNP sites | 39 | 46 | -7 |
At the same time, we can compare to variants called from reads aligned with bwa mem.
Indeed, calling variants with HPRC-corrected alignments removes 1699
SNVs (which could be false positives) and gains 3500 indels, which are the most
likely to be affected by reference bias.
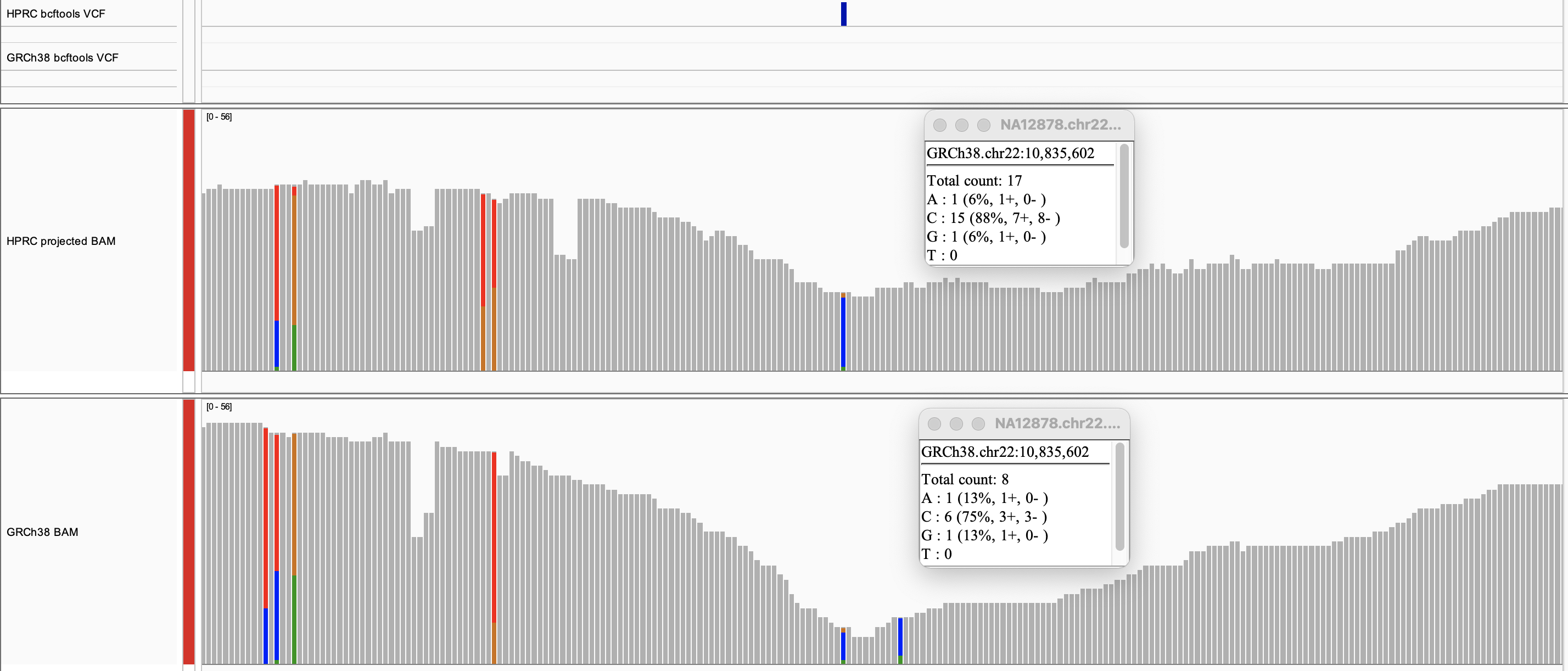
Here we see a SNV that bcftools called with HPRC but missed with GRCh38
alignments in an IGV coverage track:
First, we notice that the read depth on this SNV is slightly higher in HPRC than GRCh38.
As shown by the insert tables, HPRC enabled the recovery of 15 reads that support the
C SNV, while GRCh38 recovered only 6.
The VCF files produced by vg call also indicate which nodes represent the
variant.
Command 15
bcftools view -H /data/NA12878.chr22.hprc.vcf | lessThe ALT field indicates the source node, the node from which the graph
branches out and the sink, the node to which the graph merges again. The source
and sink nodes delimit the boundaries of a polymorphic locus. The AT field
enumerates the exact path of that variant. As an exercise, pick a variant,
subset the graph in this region with odgi, download and visualize it with
Bandage by finding the source node of a variant.