covdata is a data package for R. It provides COVID-19 related data from the following sources:
- National level case and mortality data from the European Centers for Disease Control.
- State-level case and mortality data for the United States from the COVID Tracking Project.
- State-level and county-level case and mortality data for the United States from the New York Times.
- Data from the US Centers for Disease Control's Coronavirus Disease 2019 (COVID-19)-Associated Hospitalization Surveillance Network (COVID-NET). See below for details about this network and the scope of its coverage.
- Data from Apple on relative trends in mobility in cities and countries since mid-January of 2020, based on usage of their Maps application.
- Data from Google on relative trends in mobility in regions and countries since mid-January of 2020, based on location and activity information.
- Data from the CoronaNet Research Project, providing event-based tracking of governmental policy responses to the corona virus.
The data are provided as-is. More information about collection methods, scope, limits, and possible sources of error in the data can be found in the documentation provided by their respective sources. (Follow the links above.)
Data are current through Saturday, May 23, 2020.
There are two ways to install the covdata package.
You can install covdata from GitHub with:
remotes::install_github("kjhealy/covdata")While using install_github() works just fine, it would be nicer to be able to just type install.packages("covdata") or update.packages("covdata") in the ordinary way. We can do this using Dirk Eddelbuettel's drat package. Drat provides a convenient way to make R aware of package repositories other than CRAN.
First, install drat:
if (!require("drat")) {
install.packages("drat")
library("drat")
}Then use drat to tell R about the repository where covdata is hosted:
drat::addRepo("kjhealy")You can now install covdata:
install.packages("covdata")To ensure that the covdata repository is always available, you can add the following line to your .Rprofile or .Rprofile.site file:
drat::addRepo("kjhealy")With that in place you'll be able to do install.packages("covdata") or update.packages("covdata") and have everything work as you'd expect.
Note that my drat repository only contains data packages that are not on CRAN, so you will never be in danger of grabbing the wrong version of any other package.
library(tidyverse) # Optional but strongly recommended
library(covdata)
#>
#> Attaching package: 'covdata'
#> The following objects are masked _by_ '.GlobalEnv':
#>
#> apple_mobility, cdc_deaths_by_age, cdc_deaths_by_week, cdc_hospitalizations, coronanet, covnat, covus, google_mobility, nytcovcounty, nytcovstate,
#> nytcovus
#> The following object is masked from 'package:socviz':
#>
#> %nin%
#> The following object is masked from 'package:kjhutils':
#>
#> %nin%
covnat
#> # A tibble: 18,766 x 8
#> # Groups: iso3 [209]
#> date cname iso3 cases deaths pop_2018 cu_cases cu_deaths
#> <date> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 2019-12-31 Afghanistan AFG 0 0 37172386 0 0
#> 2 2019-12-31 Algeria DZA 0 0 42228429 0 0
#> 3 2019-12-31 Armenia ARM 0 0 2951776 0 0
#> 4 2019-12-31 Australia AUS 0 0 24992369 0 0
#> 5 2019-12-31 Austria AUT 0 0 8847037 0 0
#> 6 2019-12-31 Azerbaijan AZE 0 0 9942334 0 0
#> 7 2019-12-31 Bahrain BHR 0 0 1569439 0 0
#> 8 2019-12-31 Belarus BLR 0 0 9485386 0 0
#> 9 2019-12-31 Belgium BEL 0 0 11422068 0 0
#> 10 2019-12-31 Brazil BRA 0 0 209469333 0 0
#> # … with 18,756 more rowsapple_mobility %>%
filter(region == "New York City", transportation_type == "walking")
#> # A tibble: 127 x 11
#> geo_type region transportation_type alternative_name sub_region country x2020_05_19 x2020_05_20 x2020_05_21 date index
#> <chr> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <date> <dbl>
#> 1 city New York City walking NYC New York United States 41.3 43.0 45.9 2020-01-13 100
#> 2 city New York City walking NYC New York United States 41.3 43.0 45.9 2020-01-14 96.1
#> 3 city New York City walking NYC New York United States 41.3 43.0 45.9 2020-01-15 106.
#> 4 city New York City walking NYC New York United States 41.3 43.0 45.9 2020-01-16 102.
#> 5 city New York City walking NYC New York United States 41.3 43.0 45.9 2020-01-17 117.
#> 6 city New York City walking NYC New York United States 41.3 43.0 45.9 2020-01-18 115.
#> 7 city New York City walking NYC New York United States 41.3 43.0 45.9 2020-01-19 110.
#> 8 city New York City walking NYC New York United States 41.3 43.0 45.9 2020-01-20 88.6
#> 9 city New York City walking NYC New York United States 41.3 43.0 45.9 2020-01-21 91.1
#> 10 city New York City walking NYC New York United States 41.3 43.0 45.9 2020-01-22 98.5
#> # … with 117 more rowscovus %>%
filter(measure == "positive",
date == "2020-04-27",
state == "NJ")
#> # A tibble: 1 x 5
#> date state fips measure count
#> <date> <chr> <chr> <chr> <dbl>
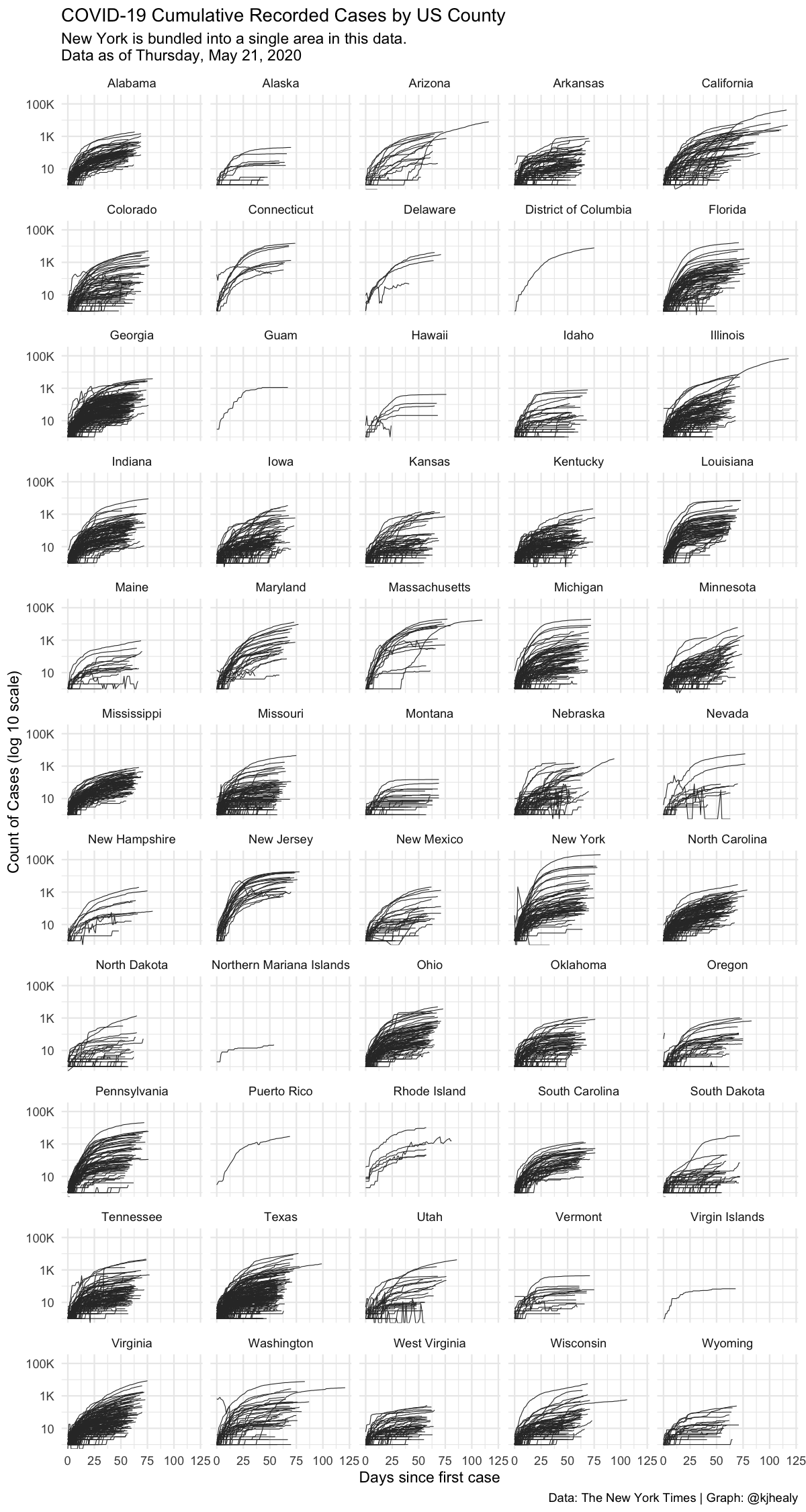
#> 1 2020-04-27 NJ 34 positive 111188nytcovcounty %>%
mutate(uniq_name = paste(county, state)) %>% # Can't use FIPS because of how the NYT bundled cities
group_by(uniq_name) %>%
mutate(days_elapsed = date - min(date)) %>%
ggplot(aes(x = days_elapsed, y = cases, group = uniq_name)) +
geom_line(size = 0.25, color = "gray20") +
scale_y_log10(labels = scales::label_number_si()) +
guides(color = FALSE) +
facet_wrap(~ state, ncol = 5) +
labs(title = "COVID-19 Cumulative Recorded Cases by US County",
subtitle = paste("New York is bundled into a single area in this data.\nData as of", format(max(nytcovcounty$date), "%A, %B %e, %Y")),
x = "Days since first case", y = "Count of Cases (log 10 scale)",
caption = "Data: The New York Times | Graph: @kjhealy") +
theme_minimal()
#> Don't know how to automatically pick scale for object of type difftime. Defaulting to continuous.
#> Warning: Transformation introduced infinite values in continuous y-axisTo learn more about the different datasets available, consult the vignettes or, equivalently, the the package website.
To cite the package use the following:
citation("covdata")
#>
#> To cite the package `covdata` in publications use:
#>
#> Kieran Healy. 2020. covdata: COVID-19 Case and Mortality Time Series. R package version 0.1.0, <http://kjhealy.github.io/covdata>.
#>
#> A BibTeX entry for LaTeX users is
#>
#> @Manual{,
#> title = {covdata: COVID-19 Case and Mortality Time Series},
#> author = {Kieran Healy},
#> year = {2020},
#> note = {R package version 0.1.0},
#> url = {http://kjhealy.github.io/covdata},
#> }Please be sure to also cite the specific data sources, as described in the documentation for each dataset.
Mask icon in hex logo by Freepik.