Last update: 2023-01-05
"In silico multiple single-subject neural tissue screening using deconvolution on pseudo-bulk RNA-seq - a prototype" (Chia et al, 2022)
This public repository showcases the prototype described in the above-mentioned thesis report.
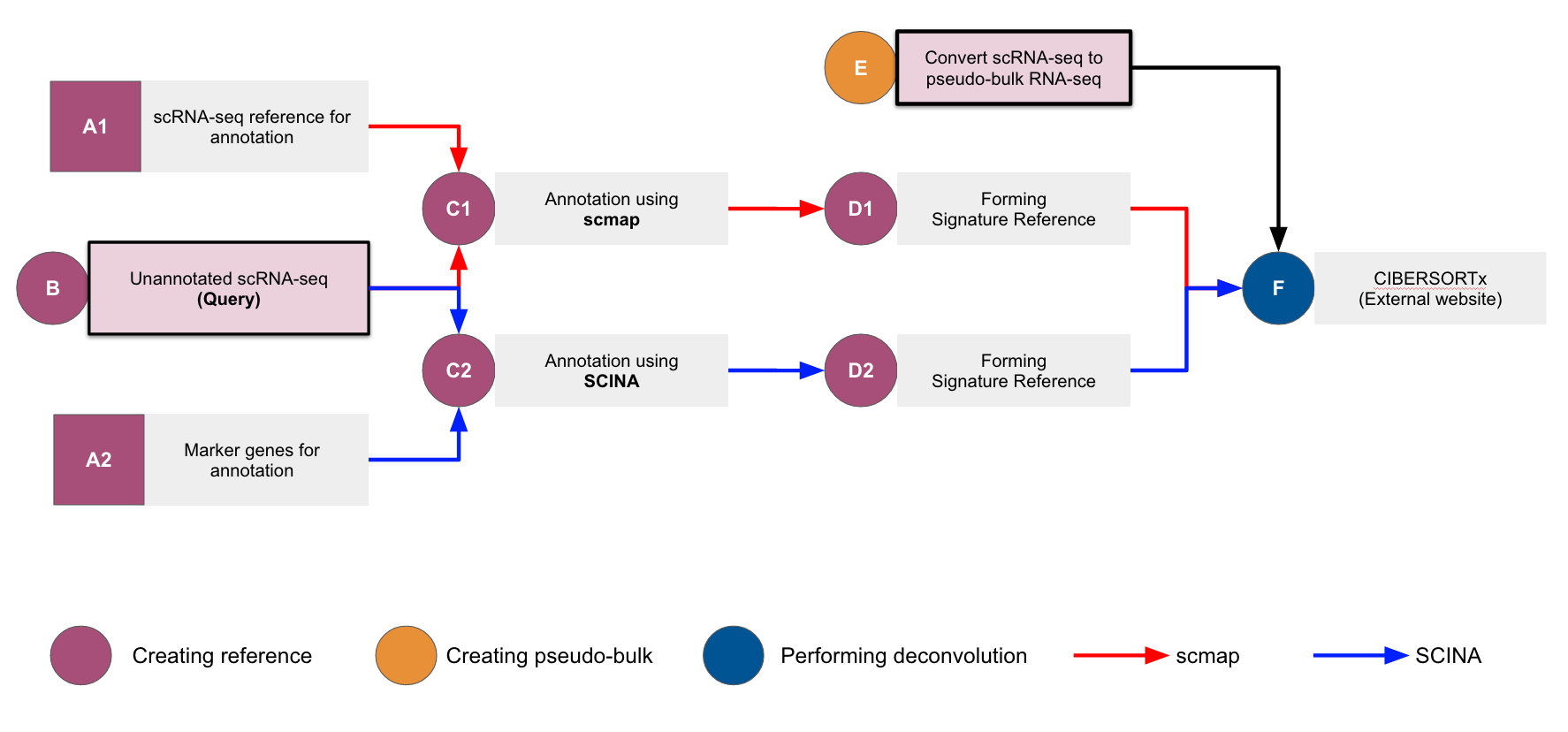
The full workflow deconvolves a bulk RNA-seq and outputs cell-type composition per sample. The deconvolution process refers to an annotated scRNA-seq (reference). Which means, the output cell-type composition is following the cell-types annotated in the input reference.
- Single-subject tissue screening - In the context of this study, this phrase means the identification of cell-type composition of a tissue sample extracted from an individual subject
- Single-cell RNA-seq (scRNA-seq) - A transcriptomics tool using RNA sequencing (RNA-seq) of individual cells
- Bulk RNA-seq - Another transcriptomics analysis taking the average expression of individual genes across cells
- Pseudo-bulk RNA-seq - Bulk RNA-seq created using an scRNA-seq dataset, commonly done via either averaging or summing the individual gene expression across all cells (reviewed in Zimmerman et al, 2021)
- Annotation - In silico (no staining required :) individual cell-type annotation based on the gene expressions of the cell
- Deconvolution - "Un-mixing" the bulk RNA-seq to cell-type composition (e.g. 49% Glutamatergic neurons, 4% GABAergic neurons, 47% Non-neuronal cells) for each tissue sample
- Annotation - a module that creates reusable references (think dictionary)
- Conversions for deconvolution - a module that converts the reference and (pseudo) bulk RNA-seq to formats acceptable in CIBERSORTx (https://cibersortx.stanford.edu/) (Newman et al, 2015)
- Reference and bulk RNA-seq should share similar biology methods and protocols
- In creating a reference using the annotation module, the input unannotated scRNA-seq must be pre-processed first
- In using CIBERSORTx (https://cibersortx.stanford.edu/), it is advised to keep the bulk RNA-seq gene expression in count values without data transformation (e.g. log-transformation).
- The tutorial notebooks were generated on a high performane computing cluster node, with these specifications: Intel(R) Xeon(R) Gold 6130 CPU @ 2.10GHz, 64 bits, and 32GB memory
- pre-processed and annotated scRNA-seq, derived from 3 tissue samples of primary somatosensory cortex of adult mouse brains [2]. This dataset is divided into 3 parts:
- Annotation-preserved scRNA-seq as a reference for the annotation process (scmap [4])
- Unannotated (where the preannotation is stripped, simulating fresh data without annotation) scRNA-seq
- Unannotated scRNA-seq which is converted into a pseudo-bulk RNA-seq via aggregation by sum
- Marker genes extracted from PanglaoDB [3] and Allen Brain Map [2]
-
Install the necessary libraries/packages listed in each Jupyter notebook and install Jupyter (https://jupyter.org/install). Alternatively, create an Anaconda environment with the provided environment.yml file.
-
Prepare 3 files (alternatively, you may download the demo files here: Finding_synapses_datasets)
- An unannotated scRNA-seq
- An annotated scRNA-seq (if you intend to use scmap to annotate the unannotated scRNA-seq)
- A bulk RNA-seq derived from 1 or more samples
- The Jupyter notebooks are labeled according to the flow chart. Start from the first notebook (A1_scmap_ReferenceForAnnotation_(Python).ipynb) or go directly to the notebook with steps of your interest.
Found a bug? Please let us know by using the Issues tab above.
Catherine Chia, Amparo Roig Adam and Alessandro Moro
Functional Genomics (FGA) Department, Center for Neurogenomics and Cognitive Research (CNCR), Vrije Universiteit Amsterdam
-
Chia, C. M., Roig Adam, A., & Moro, A. (2022). In silico multiple single-subject neural tissue screening using deconvolution on pseudo-bulk RNA-seq - a prototype. Bioinformatics and Systems Biology joint degree program. Vrije Universiteit Amsterdam and University of Amsterdam.
-
Allen Institute for Brain Science (2004). Allen Mouse Brain Atlas, Mouse Whole Cortex and Hippocampus 10x. Available from mouse.brain-map.org. Allen Institute for Brain Science (2011).
-
Franzén, O., Gan, L. M., & Björkegren, J. L. M. (2019). PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database : the journal of biological databases and curation, 2019, baz046. https://doi.org/10.1093/database/baz046
-
Kiselev, V. Y., Yiu, A., & Hemberg, M. (2018). scmap: projection of single-cell RNA-seq data across data sets. Nature methods, 15(5), 359–362. https://doi.org/10.1038/nmeth.4644
-
Zhang, Z., Luo, D., Zhong, X., Choi, J. H., Ma, Y., Wang, S., Mahrt, E., Guo, W., Stawiski, E. W., Modrusan, Z., Seshagiri, S., Kapur, P., Hon, G. C., Brugarolas, J., & Wang, T. (2019). SCINA: A Semi-Supervised Subtyping Algorithm of Single Cells and Bulk Samples. Genes, 10(7), 531. https://doi.org/10.3390/genes10070531