Code for analysing functional connectivity of cerebellar degeneration patients for manuscript Nettekoven et al., 2024.
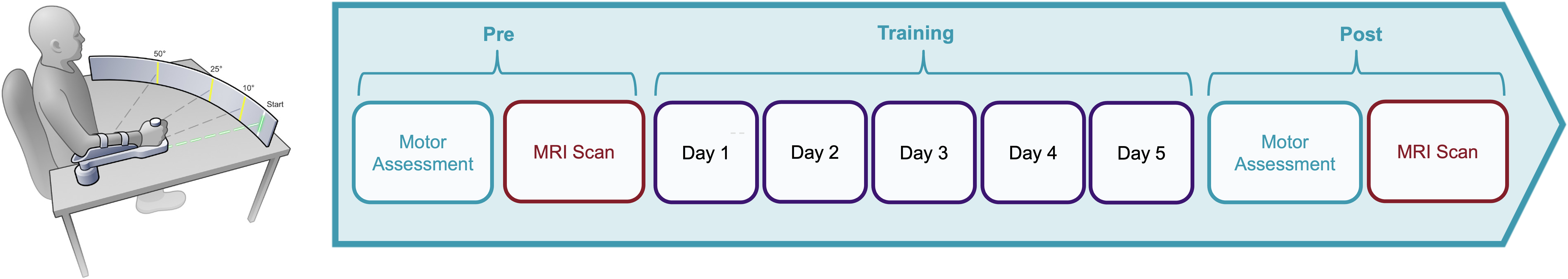
The data files available in this repository were derived from MRI scans of 40 patients diagnosed with pure cerebellar cortical degeneration and 40 age and sex-matched neurologically healthy individuals. All individuals participated a five-day motor training. On the days before and after training, participants underwent a structural MRI scan and a functional resting-state scan in addition to a motor assessment.
Functional data is missing for (subject, timepoint):
- sub-57, post
- sub-70, pre
- sub-70, post
Structural data was acquired from all subjects.
Dependencies for all code sections: see requirements.txt (run pip install -r requirements to install all required packages into your environment)
Study demographics, demographics for template generation and validation sample (Supplementary Table 1), and demographics for FIX training datasets
notebooks/demographics.ipynb
Fissure distances in template, SUIT and MNI space was calculated using:
scripts/compare_fissures.py
Fissure overlap was plotted (Fig 1B-D) and compared using:
notebooks/stats_template.ipynb
FIX performance with template registrations and standard registrations was plotted (Fig 1E) and evaluated using:
notebooks/stats_fix.ipynb
The study-specific template and associated files can be found in the DegenerationControlTemplate repository.
ROI timecourses were extracted from functional data in native space using:
scripts/seed_ts.sh
Functional connectivity between timecourses was calculated using:
scripts/seed_corr.py
Data was loaded from dataframes, normalized and brought into the correct shape for analysis using:
r/data_connectivity.R
Baseline connectivity differences were plotted (Fig 3A & 3B) using:
r/plots_connectivity.R
Connectivity change was plotted (Fig 4, 5, 6 & 7) using:
notebooks/plots_connectivity_change.ipynb
Statistical tests on functional connectivity were calculated using:
r/stats.R
Model assumptions were tested using:
r/model_assumptions.R
Connectivity results were plotted using:
r/plots_connectivity.R