This packages contains a series of functions and classes to set up and fit a Probabilistic Spatial Partition Model and sample partitionings following the the methodology introduced by Müller-Crepon, Schvitz and Cederman (2023). In short, the model is based on a simplification of continuous geographic space as a (planar) graph. The observed partitioning is encoded on the graphs’ vertices such that each vertex is a member of one and only one partition. The graphs partitioning is modeled as the result of attractive and repulsive force active on its edges which affect the probability that the vertices they connect belong to the same or to different partitions. The model allows to estimate the effects of edge-level attributes on the repulsion/attraction between vertices, thus estimating their impact on the overall partitioning of the graph.
Please refer to the original publication for all technical details beyond the summary provided below. It can be found here and as ungated version here.
Note that our companion R-package
SpatialLattice simplifies
the creation and handling of spatial graphs to facilitate the analysis
of spatial partitionings.
When using the pspm package, please cite:
Müller-Crepon, Carl, Guy Schvitz, Lars-Erik Cederman (2023). Shaping States into Nations: The Effects of Ethnic Geography on State Borders. American Journal of Political Science, conditionally accepted for publication.
We model the distribution over all possible partitionings
here, a partitioning
Potential and realized edge-level energies are in turn affected by a
constant (
The empirical goal of the PSPM is it to estimate parameters
You can directly download and install the pspm package from GitHub. Before doing so, please make sure that you have Python3 installed, which the package heavily relies on. For simplicity and as shown below, you can also install python and necessary modules from within R, using ’s and functions.
Upon installation and first usage, the package should automatically install all necessary python dependencies via the reticulate R-package. If this is not the case, the user may see an error and have install the following modules manually: scipy, networkx, numpy, abc, typing, collections.
# # Get python ready if not yet installed
# library(reticulate)
# reticulate::install_miniconda()
# reticulate::py_install(c("scipy","networkx"))
# Download pspm package
library(devtools)
install_github(repo = "carl-mc/pspm")the pspm package heavily builds on the igraph library which is used to
handle the underlying network data and allows user-friendly data
manipulation. Since random sampling plays an important role during
sampling, it is recommended to set a random seed in R and python.
pspm_set_seed() achieves just that.
# # If multiple python versions installed, point reticulate to python 3.x
# your_python_path <- '/usr/bin/python3' ## customize to your machine!
# Sys.setenv(RETICULATE_PYTHON = your_python_path)
# Packages
library(pspm)
library(igraph)
# Set seeds in R and python
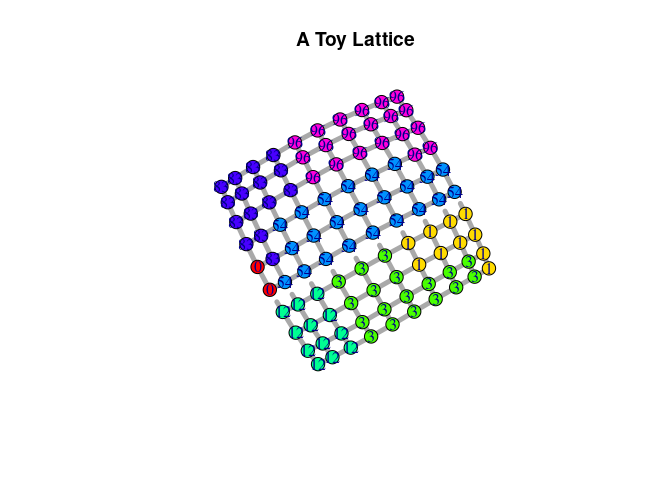
pspm_set_seed(1)To illustrate the model. we first set up a toy lattice – as a PSPM
object – with 100 nodes arranges on a 10-by-10 grid. The lattice encodes
two edge-level characteristics that separate one side of the lattice
from the other – think of a river or mountain range. The distribution of
one of these attributes is shown as dotted vs. straight edges on the
graph below. Both attributes exert repulsive forces (beta = c(2,1)).
Furthermore, the vertices are attracted to each other by a baseline
constant of -2. We sample a partitioning of the lattice with a burn-in
period of 10 rounds.
Please see the R-package
SpatialLattice for code
and examples on how to construct spatial graph data for observed spatial
partitionings.
# Make mock PSPM Object with a sampled partitioning
sl <- generate_grid_data(N_sqrd = 10, ## 10 x 10 lattice
beta0 = -2, ## Negative constant = baseline attraction between nodes
beta = c(2,1), ## Include two repulsive edge-level predictors
dep_structure = "von_neumann", ## Each node connects to 4 neighbors
burnin = 10 ## Sample with a 10 burn-in periods
)
# Print class of object
print(class(sl))## [1] "PSPM" "R6"
# Plot PSPM object
sl$plot_partitioning(edge_predictor = 1,
edge.width = 5, vertex.size = 10,
main = "A Toy Lattice")The PSPM object created above can be transformed into an igraph
object and vice-versa using the following methods:
# Transform PSPM to igraph
graph <- PSPM2igraph(sl)
# Transfers data as edge- and vertex-attributes
edge_attr_names(graph)## [1] "x1" "x2"
vertex_attr_names(graph)## [1] "X1" "Y"
# Transform igraph back to PSPM
sl.from.g <- igraph2PSPM(g = graph,
outcome_name = "Y",
edge_pred_names = c("x1", "x2"))Finally, we can fit the PSPM to estimate the effect of the edge-level
attributes on the partitioning of the lattice. The function
fit_pspm_model() provides a wrapper around lower-level functions and
classes, allowing the user to estimate the model directly from an
igraph object with a formula where the left-handside variable Y refers
to the vertex attribute that encodes vertices partition members and the
right-handside variables referring to edge-level predictors. Multiple
graphs can be passed to the function at the same time such that one
model is fit across them. Once a model is fit, bootstrap_pspm() allows
for carrying out a (parallelized) parametric bootstrap which can return
the full distribution of estimates as well as (basic and
percentile-based) confidence intervals.
## The easy way
### Estimate
m.simple <- fit_pspm_model(formula = Y ~ x1 + x2,
g_ls = list(graph),
return_pspm = TRUE)
### Bootstrap CIs
bs.simple <- bootstrap_pspm(m.simple,
n_boot_iter = 10, ## Should be > 100
burnin = 10, ## Could be higher, depending on complexite of graph and model
cl = 10L, ## Number of CPUs for parallelization
return_sims = TRUE, ## Return full distribution of estimates
ci_level = .95)## [1] "Load Learn Object on cluster"
## [1] "Run Bootstrap"
### Summary
summary(m.simple)## --------------------------------------------
## Maximum Likelihood estimation
## BFGS maximization, 31 iterations
## Return code 0: successful convergence
## Log-Likelihood: -13.44675
## 3 free parameters
## Estimates:
## Estimate Std. error t value Pr(> t)
## Constant -2.5097 0.5919 -4.240 2.23e-05 ***
## x1 1.9450 1.8374 1.059 0.290
## x2 0.4492 0.9306 0.483 0.629
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
## --------------------------------------------
print(bs.simple$ci_mat)## Constant x1 x2
## LB_Basic -3.3369735 1.6394587 -0.6487487
## UB_Basic -0.8817648 2.9760757 2.3140127
## LB_Perc -4.1375785 0.9139855 -1.4156026
## UB_Perc -1.6823697 2.2506025 1.5471588
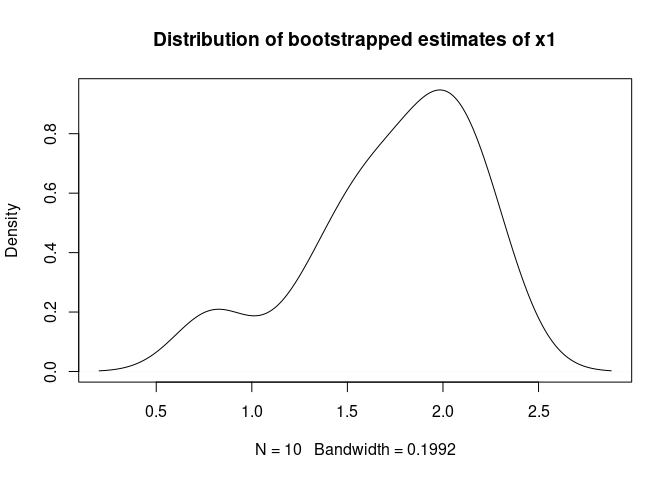
plot(density(bs.simple$beta_boot[,"x1"]),
main = "Distribution of bootstrapped estimates of x1")For completeness, the following code shows the same estimation process
using the lower-level classes and functions in the PSPM package. We
first have to transform our PSPM object into a PSPMLearn object,
which include the fit_composite_log_likelihood() method. Once the
PSPMLearn object is fitted, it can be directly bootstrapped using the
appropriate method. Note that this method does not automatically handle
missing data and other intricacies, such as keeping track of variable
names.
## The complicated way (inside the wrapper)
### Initiate PSPMLearn Object
learn_obj <- PSPMLearn$new(list(sl.from.g))
### Fit
m.compl <- learn_obj$fit_composite_log_likelihood(beta_init = c(0,0,0))
### Bootstrap
bs.compl <- learn_obj$par_bootstrap_composite_log_likelihood(n_boot_iter = 10, burnin = 10,
cl = 10L, return_sims = FALSE, ci_level = .95)## [1] "Load Learn Object on cluster"
## [1] "Run Bootstrap"
### Summary
summary(m.compl)## --------------------------------------------
## Maximum Likelihood estimation
## BFGS maximization, 31 iterations
## Return code 0: successful convergence
## Log-Likelihood: -13.44675
## 3 free parameters
## Estimates:
## Estimate Std. error t value Pr(> t)
## [1,] -2.5097 0.5919 -4.240 2.23e-05 ***
## [2,] 1.9450 1.8374 1.059 0.290
## [3,] 0.4492 0.9306 0.483 0.629
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
## --------------------------------------------
print(bs.compl)## Constant x1 x2
## LB_Basic -3.3369735 1.6394587 -0.6487487
## UB_Basic -0.8817648 2.9760757 2.3140127
## LB_Perc -4.1375785 0.9139855 -1.4156026
## UB_Perc -1.6823697 2.2506025 1.5471588
Finally, models fitted with fit_pspm_model() and the
(maxLik)[https://cran.r-project.org/web/packages/maxLik/index.html]
methods used under the hood can be printed as text, html, or latex files
using a slight extension of the
(texreg)[https://cran.r-project.org/web/packages/texreg/texreg.pdf]
package.
# Integration with texreg
pspm2table(list(m.simple),
bootci = list(bs.simple$ci_mat), boottype = "percentile",
type = "text", add.stats = c("Edges" = "N_edges", "Vertices" = "N"))##
## ==============================
## Model 1
## ------------------------------
## Constant -2.51 *
## [-4.14; -1.68]
## x1 1.95 *
## [ 0.91; 2.25]
## x2 0.45
## [-1.42; 1.55]
## ------------------------------
## Edges 180
## Vertices 100
## Log-Likelihood -13.45
## Num. obs. 100
## ==============================
## * 0 outside the confidence interval.
We are very grateful for any bug reports, feedback, questions, or contributions to this package. Please report any issues here or write to c.a.muller-crepon [at] lse.ac.uk .