version: 1.0
A Python project that allows you to analyse proteomics and clinical data, and integrate and mine knowledge from multiple biomedical databases widely used nowadays.
- Documentation: https://CKG.readthedocs.io
- GitHub: https://github.com/MannLabs/CKG
- Free and open source software: MIT license
- Reference: https://www.biorxiv.org/content/10.1101/2020.05.09.084897v1
- Graph Database dump file and additional relationships: https://data.mendeley.com/datasets/mrcf7f4tc2/1
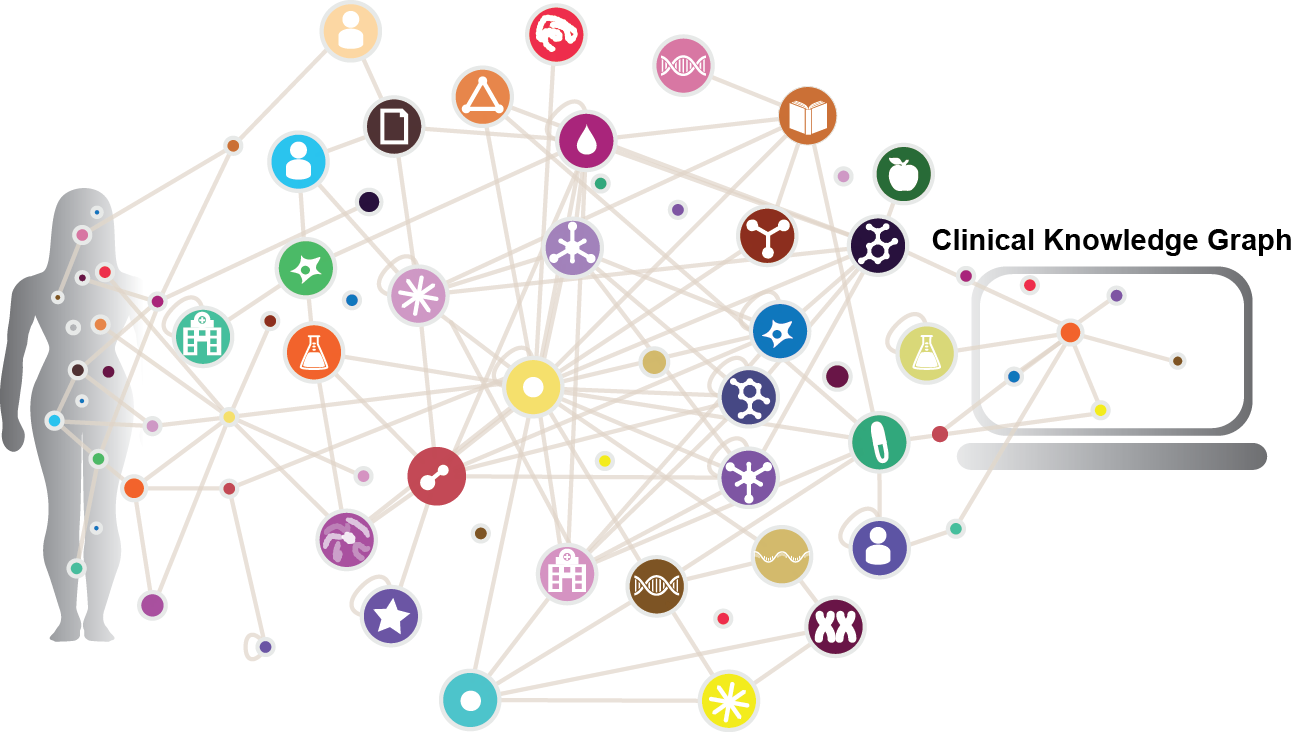
The promise of precision medicine is to deliver personalized treatment based on the unique physiology of each patient. This concept was fueled by the genomic revolution, but it is now evident that integrating other types of omics data, like proteomics, into the clinical decision-making process will be essential to accomplish precision medicine goals. However, quantity and diversity of biomedical data, and the spread of clinically relevant knowledge across myriad biomedical databases and publications makes this exceptionally difficult. To address this, we developed the Clinical Knowledge Graph (CKG), an open source platform currently comprised of more than 16 million nodes and 220 million relationships to represent relevant experimental data, public databases and the literature. The CKG also incorporates the latest statistical and machine learning algorithms, drastically accelerating analysis and interpretation of typical proteomics workflows. We use several biomarker studies to illustrate how the CKG may support, enrich and accelerate clinical decision-making.
Installation requires >= 80 GB of disk space. See details here.
The setting up of the CKG includes several steps and might take a few hours (if you are building the database from scratch). However, we have prepared documentation and manuals that will guide through every step. To get a copy of the GitHub repository on your local machine, please open a terminal windown and run:
$ git clone https://github.com/MannLabs/CKG.gitThis will create a new folder named "CKG" on your current location. To access the documentation, use the ReadTheDocs link above, or open the html version stored in the CKG folder CKG/docs/build/html/index.html. After this, follow the instructions in "First Steps" and "Getting Started".
Warning
If git is not installed in your machine, please follow this tutorial to install it.
- Cross-platform: Mac, and Linux are officially supported. Instructions for Windows exist.
- Docker container runs all neccessary steps to setup the CKG.
This resource is intended for research purposes and must not substitute a doctor’s medical judgement or healthcare professional advice.
The databases provided within the Clinical Knowledge Graph (CKG) have their own licenses and the use of CKG still requires compliance with these data use restrictions. Please, visit the data sources directly for more information: