FastSegFormer: A knowledge distillation-based method for real-time semantic segmentation of surface defects in navel oranges
This is the official repository for our work: FastSegFormer(PDF)
News
This work was accepted for publication in the journal Computers and Electronics in Agriculture on December 29, 2023.
Highlights
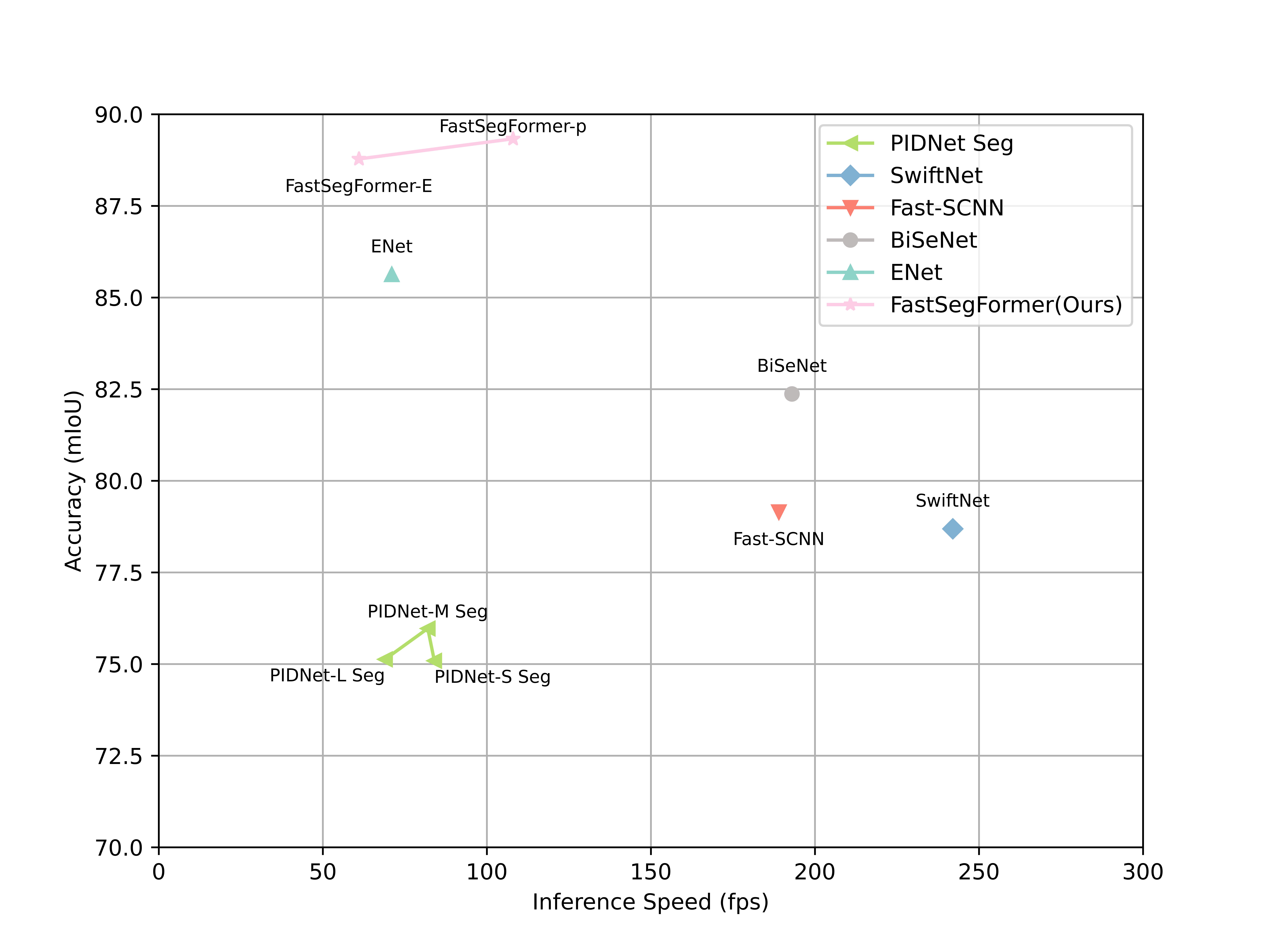
- Performance of different models on navel orange dataset (test set) against their detection speed on RTX3060:
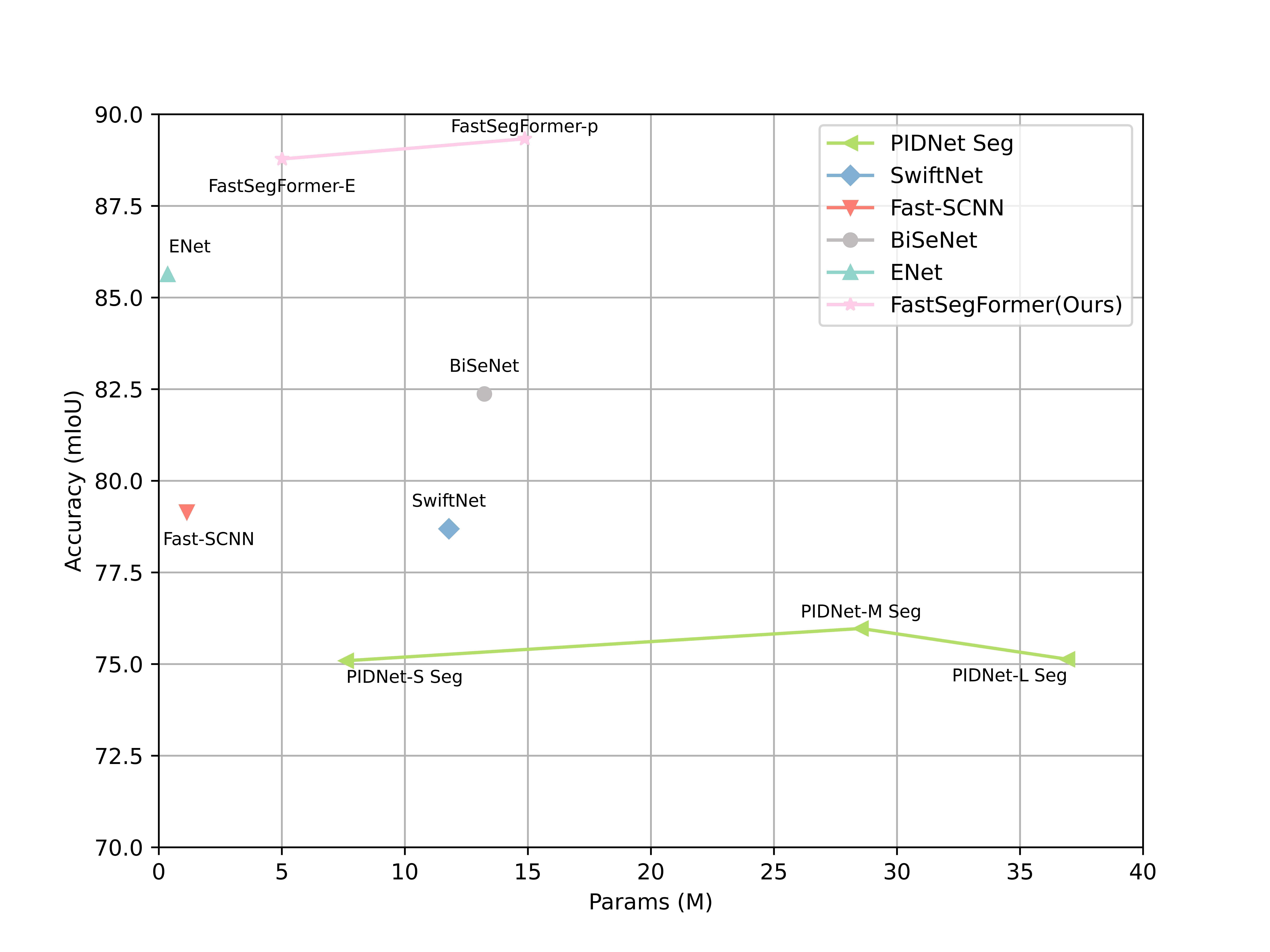
- Performance of different models on navel orange dataset (test set) against their parameters:
Updates
- The training and testing codes are available here.(April/25/2023)
- Create PyQT interface for navel orange defect segmentation. (May/10/2023)
- Produce 30 frames of navel orange assembly line simulation video. (May/13/2023)
- Add yolov8n-seg and yolov8-seg instance segmentation training, test, and prediction results.Jump to(December/10/2023)
Demos
- Some demos of the segmentation performance of our proposed FastSegFormer:Original image(left) and Label image(middle) and FastSegFormer-P(right). The original image contains enhanced image.
- A demo of Navel Orange Video Segmentation:Original video(left) and detection video(right). The actual detection video reaches 45~55 fps by weighted half-precision (FP16) quantization technique and multi-thread processing technique.(The actual video detection is the total latency of pre-processing, inference and post-processing of the image). Navel orange defect picture and video detection UI is available at FastSegFormer-pyqt.


Navel orange simulation line detection video
Overview
-
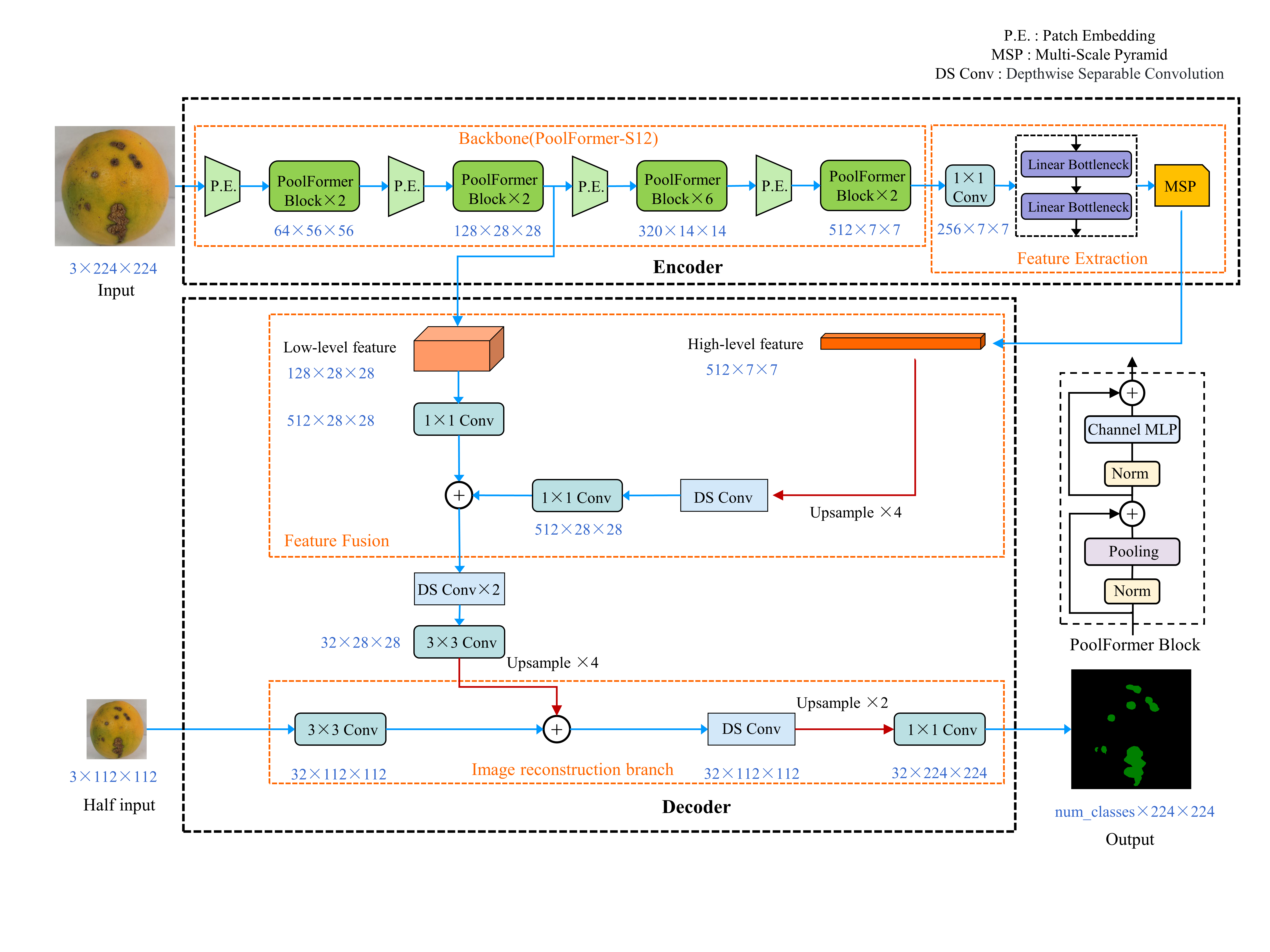
An overview of the architecture of our proposed FastSegFormer-P. The architecture of FastSegFormer-E is derived from FastSegFormer-P replacing the backbone network EfficientFormerV2-S0.
-
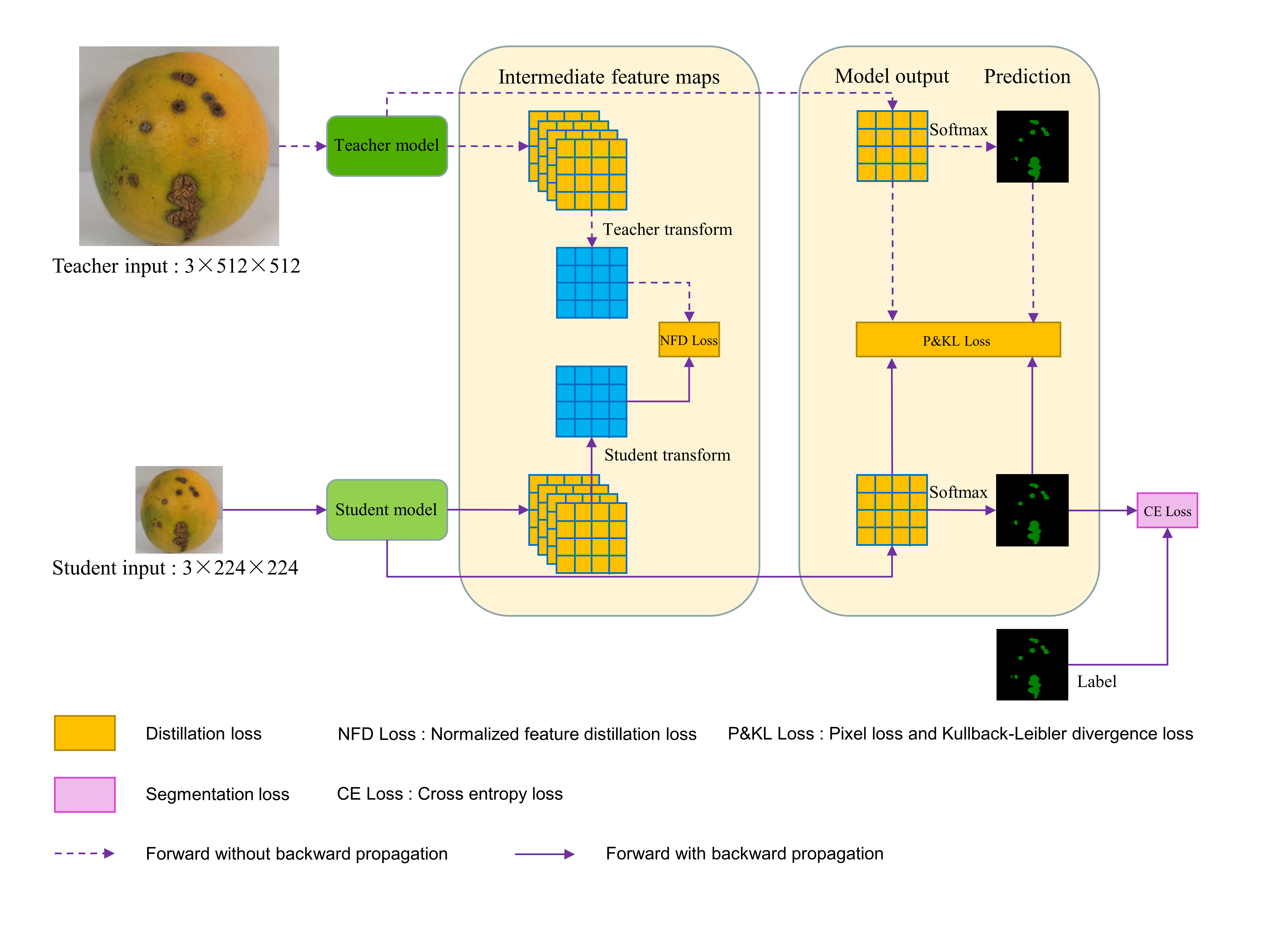
An overview of the proposed multi-resolution knowledge distillation.(To solve the problem that the size and number of channels of the teacher network and student network feature maps are different:the teacher network's feature maps are down-sampled by bilinear interpolation, and the student network's feature maps are convolved point-by-point to increase the number of channels)
P&KL loss:
Where
NFD loss:
Where
where
Models
- Pretrained backbone network:
| Model(ImageNet-1K) | Input size | ckpt |
|---|---|---|
| EfficientFormerV2-S0 | download | |
| EfficientFormerV2-S1 | download | |
| PoolFormer-S12 | download | |
| PoolFormer-S24 | download | |
| PoolFormer-S36 | download | |
| PIDNet-S | download | |
| PIDNet-M | download | |
| PIDNet-L | download |
- Teacher network:
| Model | Input size | mIoU(%) | mPA(%) | Params | GFLOPs | ckpt |
|---|---|---|---|---|---|---|
| Swin-T-Att-UNet | 90.53 | 94.65 | 49.21M | 77.80 | download |
- FastSegFormer after fine-tuning and knowledge distillation:
| Model | Input size | mIoU(%) | mPA(%) | Params | GFLOPs | RTX3060(FPS) | RTX3050Ti(FPS) | ckpt | onnx |
|---|---|---|---|---|---|---|---|---|---|
| FastSegFormer-E | 88.78 | 93.33 | 5.01M | 0.80 | 61 | 54 | download | download | |
| FastSegFormer-P | 89.33 | 93.78 | 14.87M | 2.70 | 108 | 93 | download | download |
Ablation study
You can see all results and process of our experiment in logs dir, which include ablation study and comparison
with other lightweight models.
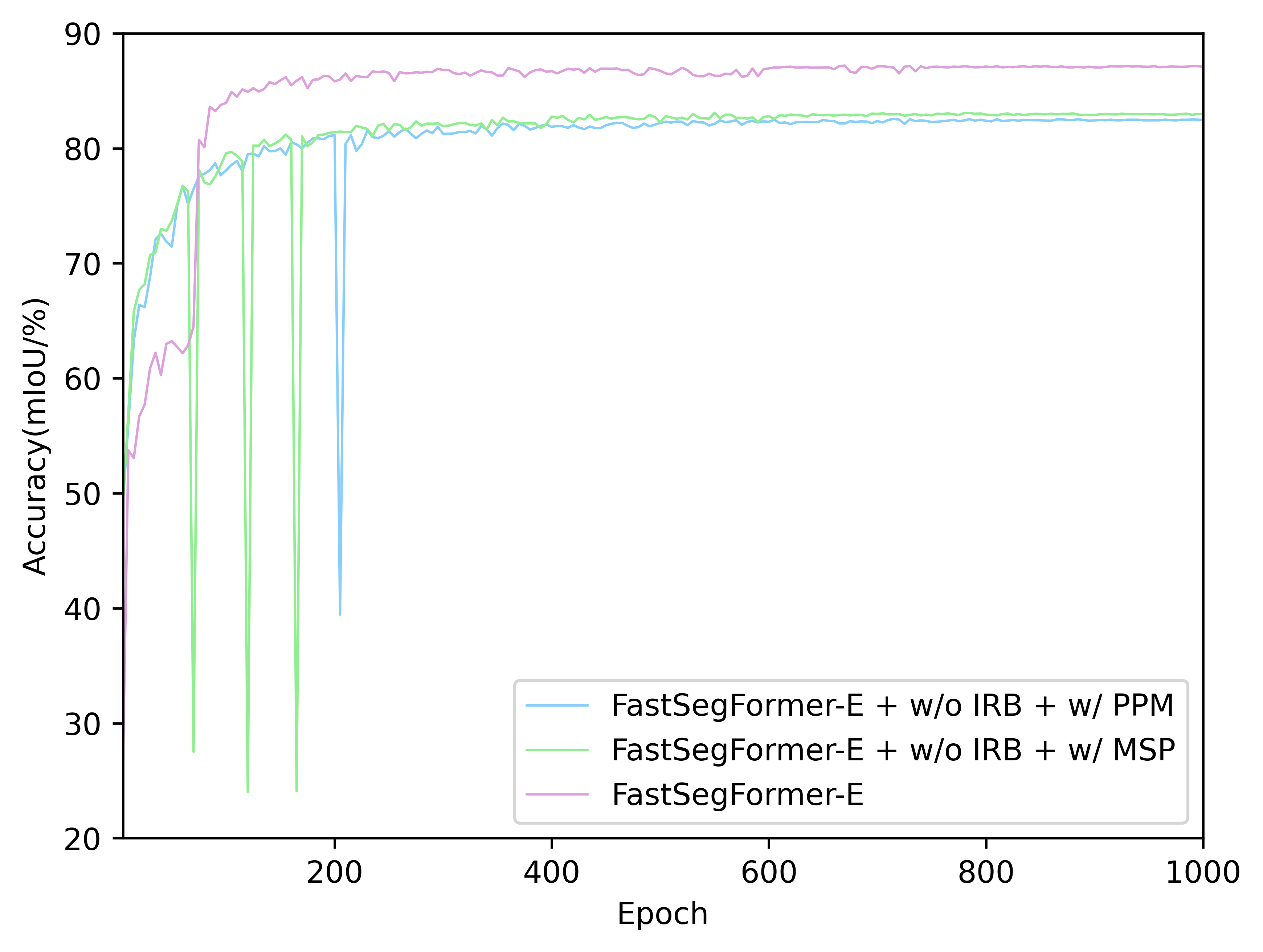
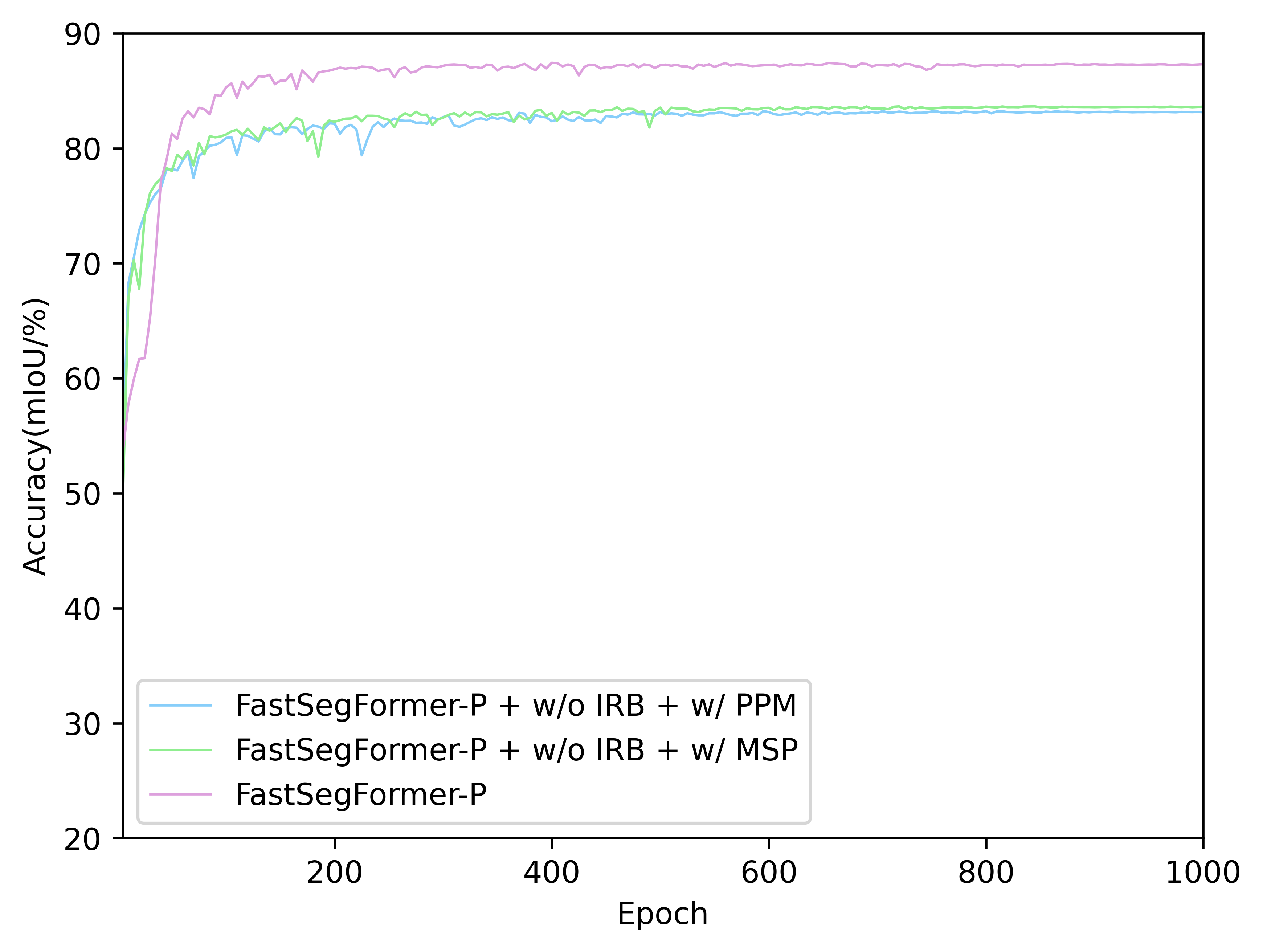
- The Acc.(mIoU) of FastSegFormer models with different network structure(PPM, MSP and Image reconstruction branch) on validation set:
- Knowledge distillation(KD) and fine-tuning(†):
| Model | mIoU(%) | mPA(%) | mPrecision(%) | Params | GFLOPs |
|---|---|---|---|---|---|
| FastSegFormer-E | 86.51 | 91.63 | 93.50 | 5.01 | 0.80 |
| FastSegFormer-E w/ |
87.24 | 92.20 | 93.82 | 5.01 | 0.80 |
| FastSegFormer-E w/ |
87.38 | 92.35 | 93.83 | 5.01 | 0.80 |
| FastSegFormer-E† | 88.49 | 93.16 | 94.32 | 5.01 | 0.80 |
| FastSegFormer-E w/ |
88.68 | 92.97 | 94.75 | 5.01 | 0.80 |
| FastSegFormer-E w/ |
88.78 | 93.33 | 94.48 | 5.01 | 0.80 |
| FastSegFormer-P | 84.15 | 89.44 | 92.84 | 14.87 | 2.70 |
| FastSegFormer-P w/ |
84.77 | 90.12 | 92.91 | 14.87 | 2.70 |
| FastSegFormer-P w/ |
85.43 | 90.64 | 93.20 | 14.87 | 2.70 |
| FastSegFormer-P† | 88.57 | 93.15 | 94.42 | 14.87 | 2.70 |
| FastSegFormer-P w/ |
88.94 | 93.25 | 94.77 | 14.87 | 2.70 |
| FastSegFormer-P w/ |
89.33 | 93.78 | 94.68 | 14.87 | 2.70 |
Environment
This implementation is based on unet-pytorch. The detection speed(FPS) is tested on single RTX3060 and on single RTX3050Ti.
- Hardware Configuration: A graphics card with 12G graphics memory is a must, as our knowledge distillation method will take up a lot of video memory during training.(When the batch size of knowledge distillation is 6, the graphics memory occupies 11.8G) Of course you can also skip the distillation method, which will take up a very low amount of graphics memory.
- Basic environment configuration: Our code currently only supports single card training. Our training environment :Python 3.9, Pytorch 1.12.1, CUDA 11.6.
Usage
Prepare the dataset
We only provide 1448 navel orange defect dataset in VOC format, if you want to extend the dataset, you can use Imgaug for segmentation maps and masks for data enhancement.
- Download the Orange_Navel_1.5k dataset and unzip them in
data/Orange_Navel_1.5kdir. - The dataset we provide has been randomly partitioned according to the training validation test 6:2:2. You
can re-randomize or re-randomize after changing the ratio using
voc_annotation.py.
Train
- Download the source code zip or clone the project:
$ git clone https://github.com/caixiongjiang/FastSegFormer- Go to the root directory of project and download the required packages:
$ conda activate 'your anaconda environment'
$ pip install -r requirements.txt- Download the ImageNet pretrained models and put them into
model_datadir.
Thanks to three repositories EfficientFormer, poolformer and PIDNet, We provide the pretraining weights of EfficientFormerV2, PoolFormer and PIDNet on ImageNet-1K.
- Modify the parameters of
train.py. For example, train FastSegFormer-P(fin-tuning):
backbone = "poolformer_s12"
pretrained = False
model_path = "model_data/poolformer_s12.pth"
input_shape = [224, 224]- Train the FastSegFormer-P model on Orange_Navel_1.5k with batch size of 32.
python train.pyKnowledge distillation(KD) train
You can train with KD or KD + fine-tuning.
-
For example, train with KD + fine-tuning, download the ImageNet pretrained model's checkpoint and download the teacher network's checkpoint(Swin-T-Att-UNet). Then put them into
model_datadir. -
Modify the parameters of
train_distillation.py. For example, train FastSegFormer-P(KD + fine-tuning):
t_backbone = "swin_T_224"
s_backbone = "poolformer_s12"
pretrained = False
t_model_path = "model_data/teacher_Swin_T_Att_Unet_input_512.pth"
s_model_path = "model_data/poolformer_s12.pth" # if s_model_path = "": from scratch else fine-tuning
Init_Epoch = 0
Freeze_Epoch = 50
Freeze_batch_size = 6
UnFreeze_Epoch = 1000
Unfreeze_batch_size = 6
Freeze_Train = True
Init_lr = 1e-5
Min_lr = Init_lr * 0.01- Train the FastSegFormer-P model using fine-tuning and KD on
Orange_Navel_1.5kwith batch size of 6.
python train_distillation.pyEvaluation of segmentation
- For example, download the fine-tuning models FastSegFormer-P for Orange_Navel_1.5k and put them into
logs/FastSegFormer-Pdir. - modify the parameters of
unet.py. For example, evaluate FastSegFormer-P(fine-tuning):
_defaults = {
"model_path" : 'logs/FastSegFormer-P.pth',
"num_classes" : 3 + 1,
"backbone" : "poolformer_s12",
"input_shape" : [224, 224],
"mix_type" : 1,
"cuda" : True, # if backbone = efficientFormerV2, cuda should be False
}
def generate(self, onnx=False):
self.net = FastSegFormer(num_classes=self.num_classes, pretrained=False, backbone=self.backbone, Pyramid="multiscale", cnn_branch=True)- Evaluate the test set on Navel_Orange_1.5k and the result will be in the
miou_outdir.
python get_miou.pypredict
Same as the evaluation session, firstly modify the parameters of unet.py, and then run predict.py:
python predict.py
# Generate tips and input the image dir
Input image filename:'your image dir'Evaluation of deployment
- Detection speed(FPS):
python speeding.py- Params and GFLOPs:
python model_flop_params.pyCitation
if you this implementation is useful for your work, please cite our paper:
Acknowledgement
- This implementation is based on unet-pytorch.
- FPS measurement code is borrowed from FANet.
- The backbone network checkpoints is download from EfficientFormer, poolformer and PIDNet.