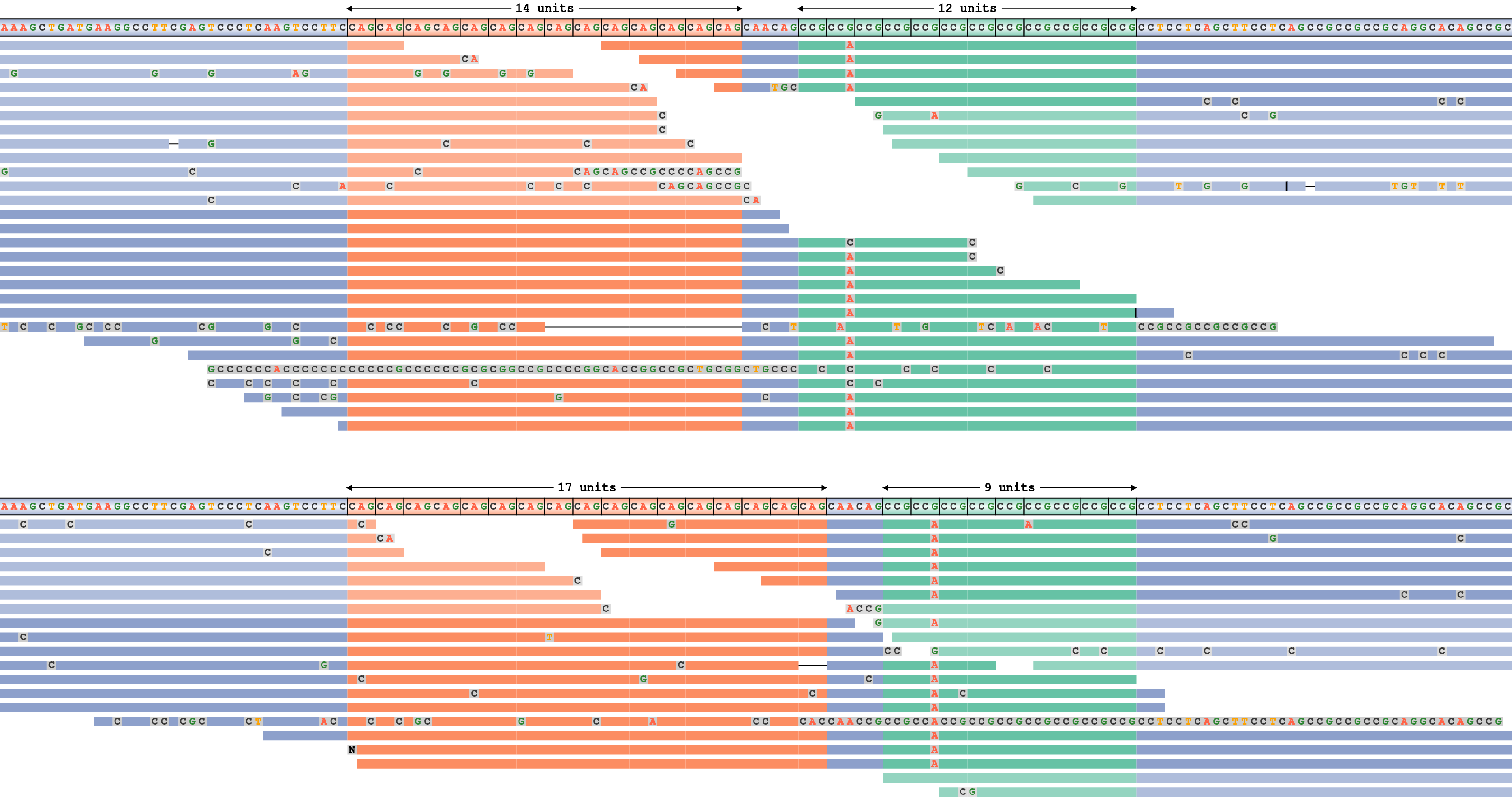
Repeat Expansion Viewer (REViewer)
REViewer is a tool for visualizing alignments of reads in regions containing tandem repeats. REViewer requires a BAMlet with graph-realigned reads generated by ExpansionHunter and the corresponding variant catalog.
License
REViewer is provided under the terms and conditions of the GPLv3 license. It relies on several third party packages provided under other open source licenses, please see COPYRIGHT.txt for additional details.
Installation
The simplest way of obtaining REViewer is by downloading a Linux binary corresponding to the latest release from the Releases page. The link to the binary is located in the Assets section.
REViewer can also be built from source with CMake.
cd REViewer
mkdir build; cd build
cmake ..; makeUsage
REViewer requires output files generated by ExpansionHunter v3.0.0 or above along with the matching variant catalog file and reference genome.
REViewer \
--reads <BAMlet generated by ExpansionHunter> \
--vcf <VCF file generated by ExpansionHunter> \
--reference <FASTA file with reference genome> \
--catalog <Variant catalog> \
--locus <Locus to analyze> \
--output-prefix <Prefix for the output files>Note that the BAMlet generated by ExpansionHunter (--reads parameter) must be
sorted and indexed.
Introductory guides
- A blog post describing the method
- Examples of read pileups corresponding to correctly and incorrectly genotyped repeats.
- You can use the files under
/reviewer/tests/inputs/to test REViewer on your own machine. (Don't use the outputs from the ExpansionHunter repository example; it contains variant locus features that REViewer does not support, and will crash REViewer.)
Reference documentation
Companion tools
-
FlipBook is an image server for REViewer developed by Ben Weisburd. It provides a convenient way to inspect large quantities of read pileups.
-
Review BAMs is script that allows applying REViewer to a regular BAM file (by running ExpansionHunter in the background). It was developed by Andreas Halman.
Citation
- Dolzhenko E, Weisburd B, Garikano K, Rajan Babu IS, and colleagues, REViewer: Haplotype-resolved visualization of read alignments in and around tandem repeats, bioRxiv, 2021