In the field of computational chemistry, the ability to quickly and easily view molecules is incredibly important. Currently, many research groups, such as the Mobley Lab at UC Irvine, use Slack for communication purposes. When they share molecule files, they are required to download the file and open it in a local viewer. This greatly hampers the efficiency of communication; furthermore, it makes it nearly impossible to view molecules on the go. MOVES (MOlecule ViEwer for Slack) is a Slack app that solves this problem. By interfacing with 3DMol.js, MOVES enables users to view molecules without the need for any external viewer.
In terms of implementation, MOVES is a server using Express.js. It currently runs on Heroku, with an Amazon S3 bucket used for storage.
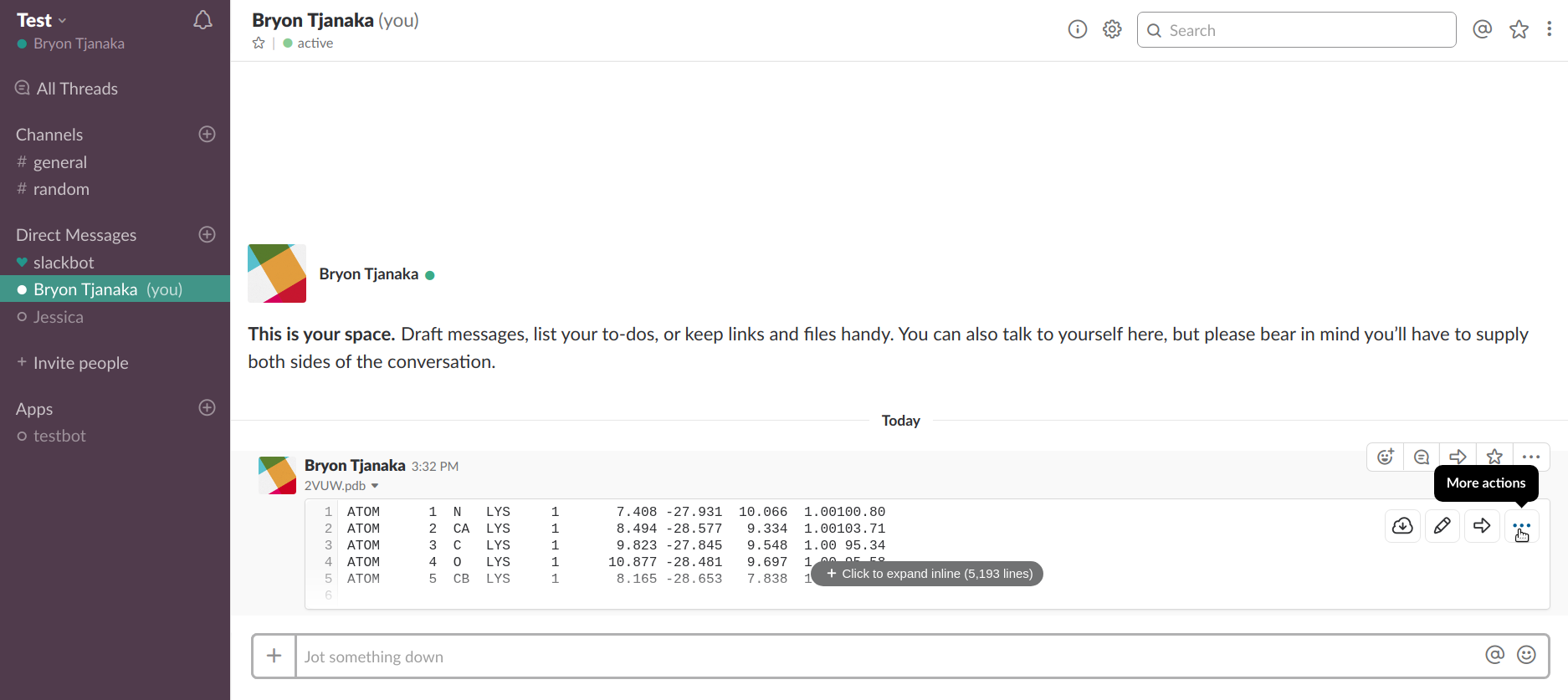
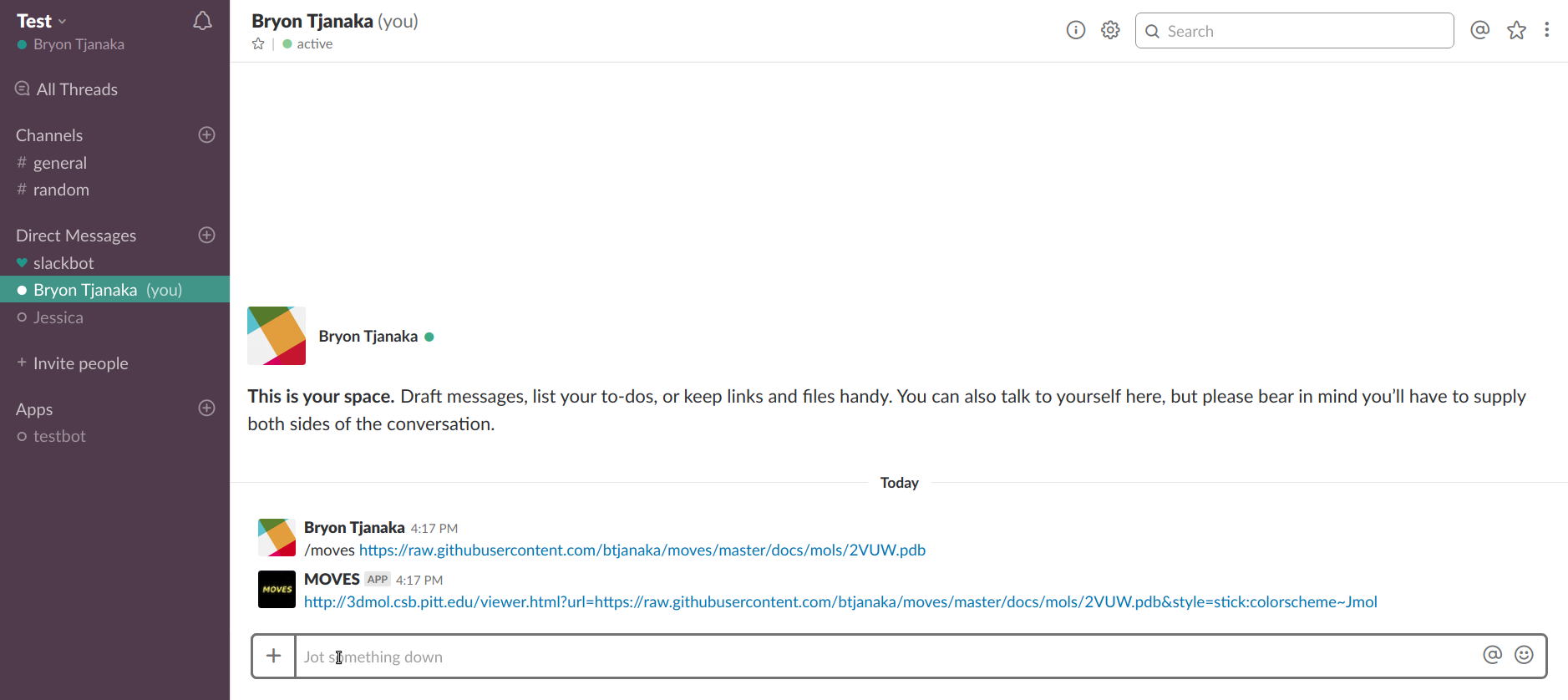
MOVES is invoked as a slash command within Slack:
/moves [file URL]
The file URL can be a link to:
- A molecule file on the team's Slack*
- A molecule file on some external link
After the file URL is passed via the slash command, MOVES will return a link where the file may be viewed using 3DMol.js, or an appropriate error message.
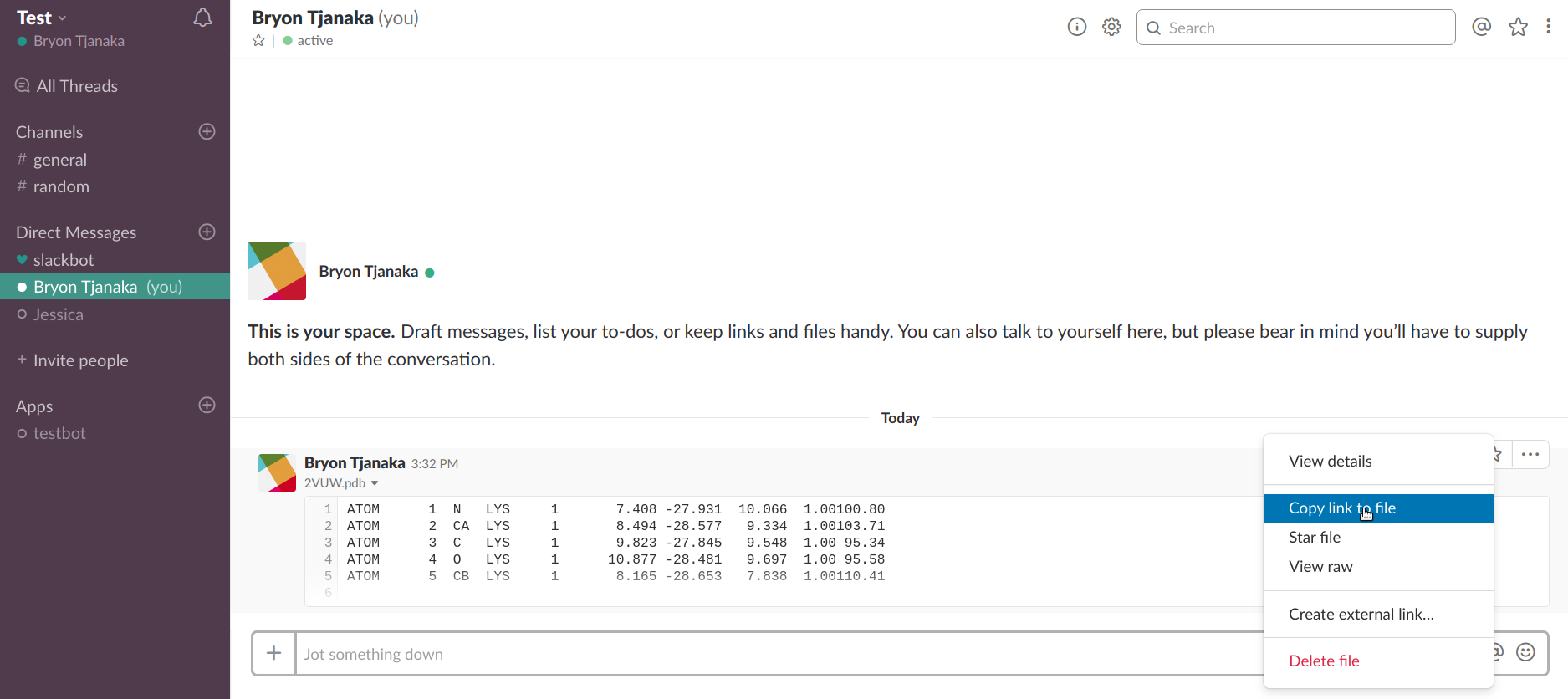
* This must be a link to a file, not a link to a message with a file. You can obtain this link by hovering over the top right corner of the preview, clicking on the three dots that appear, and clicking "Copy link to file". See the Slack tutorial for more info.
- Slack may give a timeout error for MOVES. This happens becaues the MOVES server may be inactive (It runs on a free account on Heroku, which puts the server to sleep after 30 minutes of inactivity). If this occurs, wait a few seconds and try again.
- If the link is a Slack file link, MOVES may give an error if:
- The Slack file is not one of the supported molecule file types
- The Slack file is not on the correct Slack (i.e. MOVES cannot access it)
- If the link is an external link or a Slack link (but not to a file), 3DMol.js may give a blank screen if the file does not exist or cannot be accessed.
- If the link is not a URL, an appropriate message is returned.
When given a Slack file link, MOVES downloads the file and stores it. There are two important implications associated with this:
- Slack file links are "public." Anyone with a link to the molecule file can download it. If you do not mind this, there is nothing to worry about; however, if you are developing a top secret molecule, MOVES may not be the best option.
- Slack file links may not work after 24 hours. For every 100 files it receives, the server does a sweep to remove any files that are over 24 hours old, so your file may not be available anymore. Since there is still not much traffic, this is unlikely to affect you for now, but if it does, simply send MOVES the link again on Slack.
The filetypes MOVES supports are based entirely on the ones that 3DMol.js
supports. As of writing, these filetypes are pdb, sdf, mol2, xyz, and
cube.
Currently there appears to be a bug where MOVES erroneously tells users that the Slack file they gave may be on another team's Slack. I am unsure why this happens; all I know is that it has something to do with MOVES failing to access these files.
Here are several tutorials for using MOVES to view molecule files.
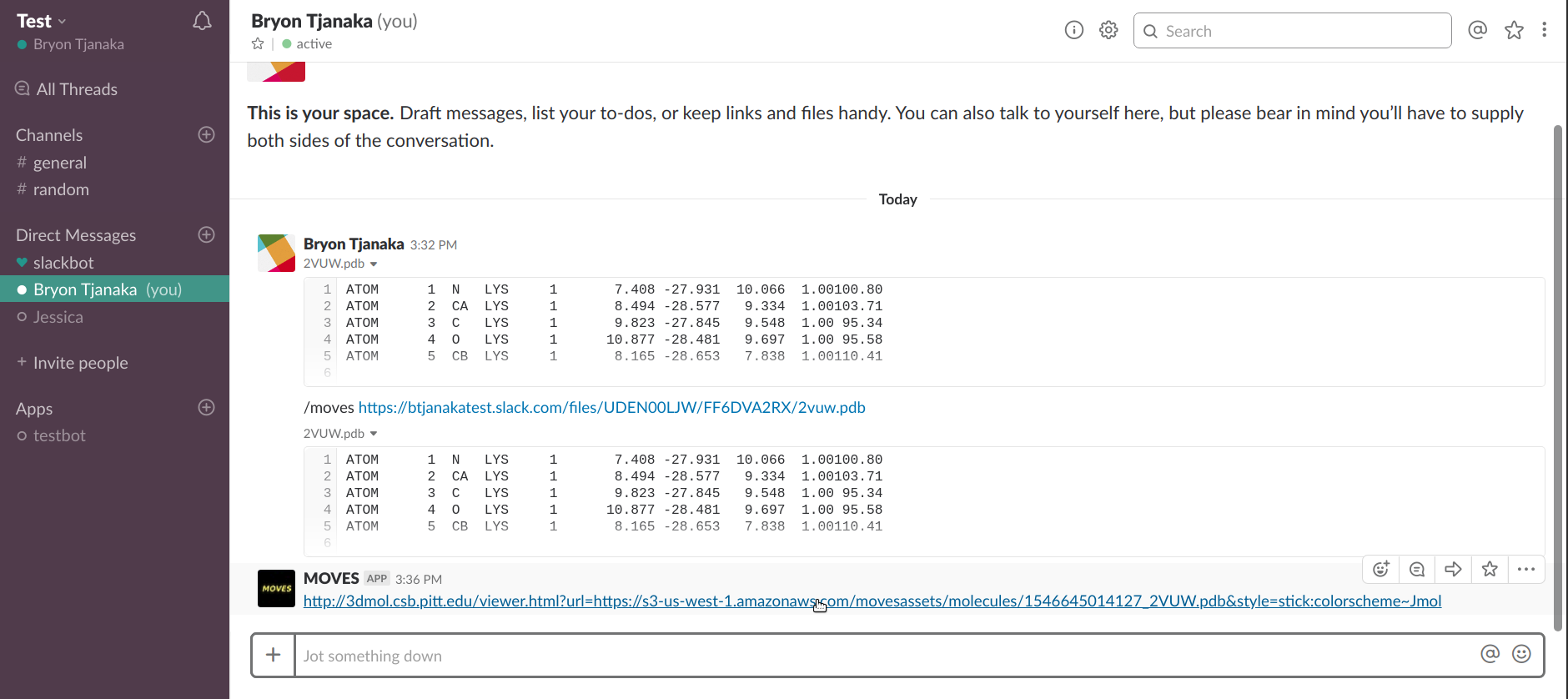
To view a Slack file, first upload the file.
Once the file is uploaded, hover over the top right corner and three dots will appear.
Click on the three dots and then "Copy link to file".
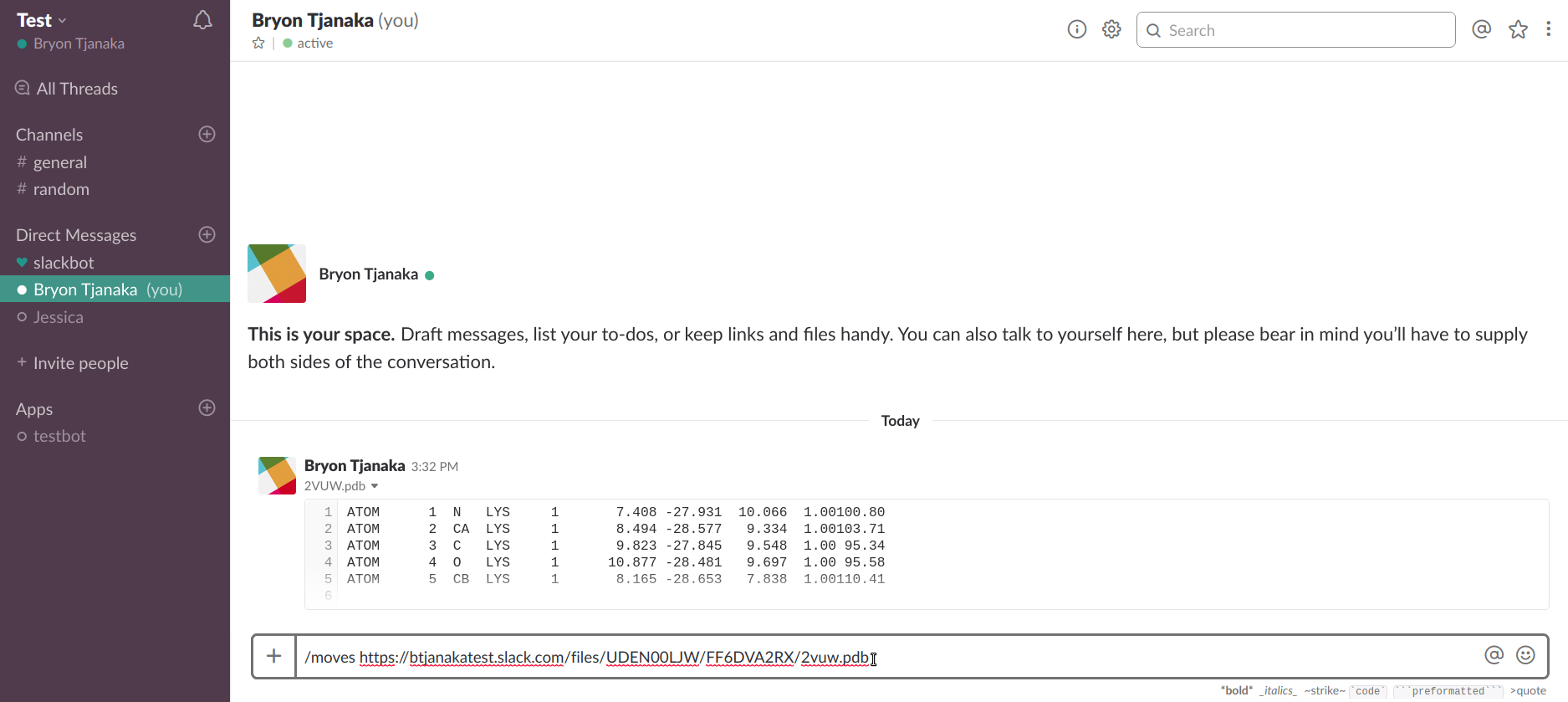
Enter the command "/moves" and paste the link.
Send the message and you should receive a link from MOVES. Click on the link to view the molecule!
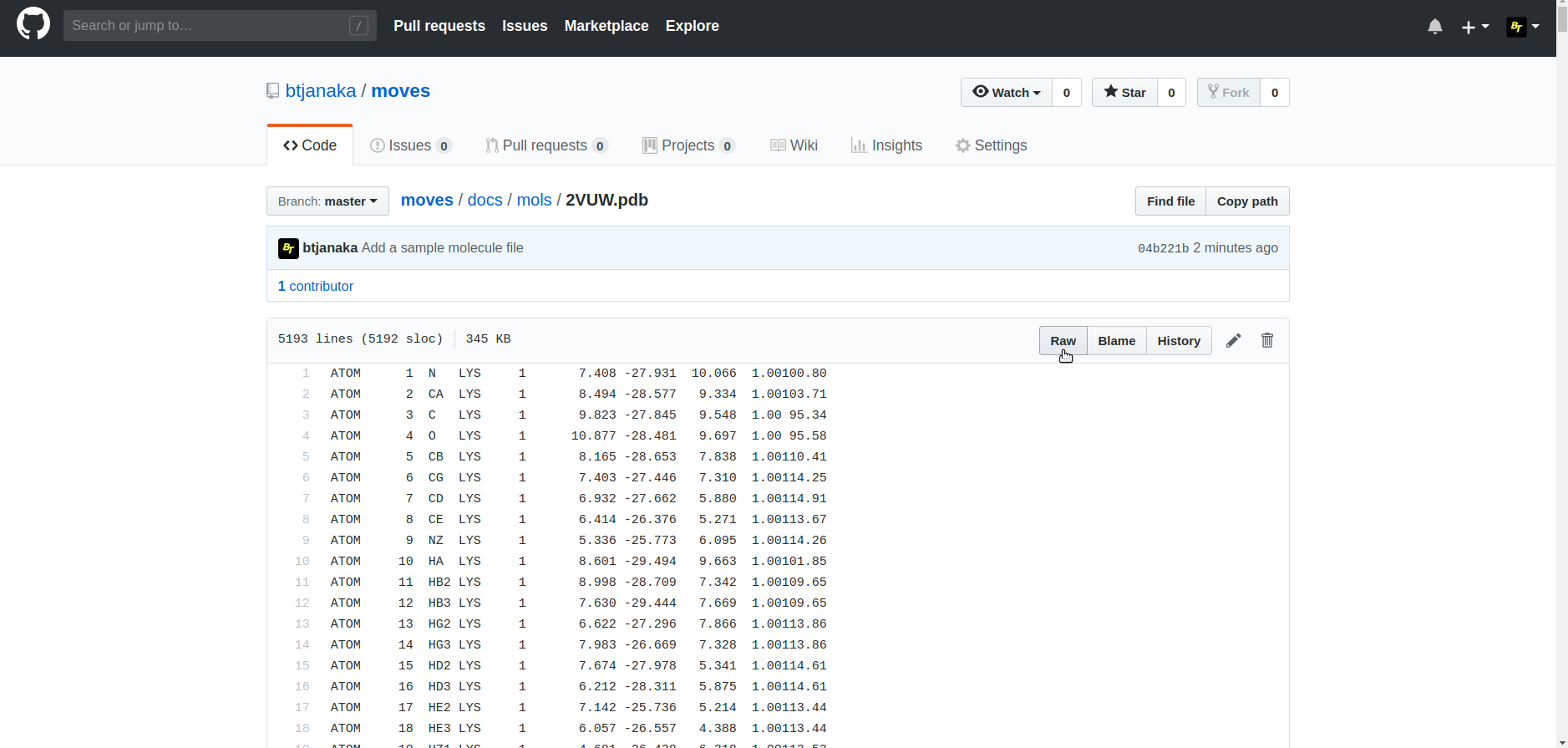
For Github molecule files, first navigate to the appropriate page on Github. Then, select the "raw" option for viewing the file.
After choosing the raw version, you might see a webpage like this. Copy the URL of the page.
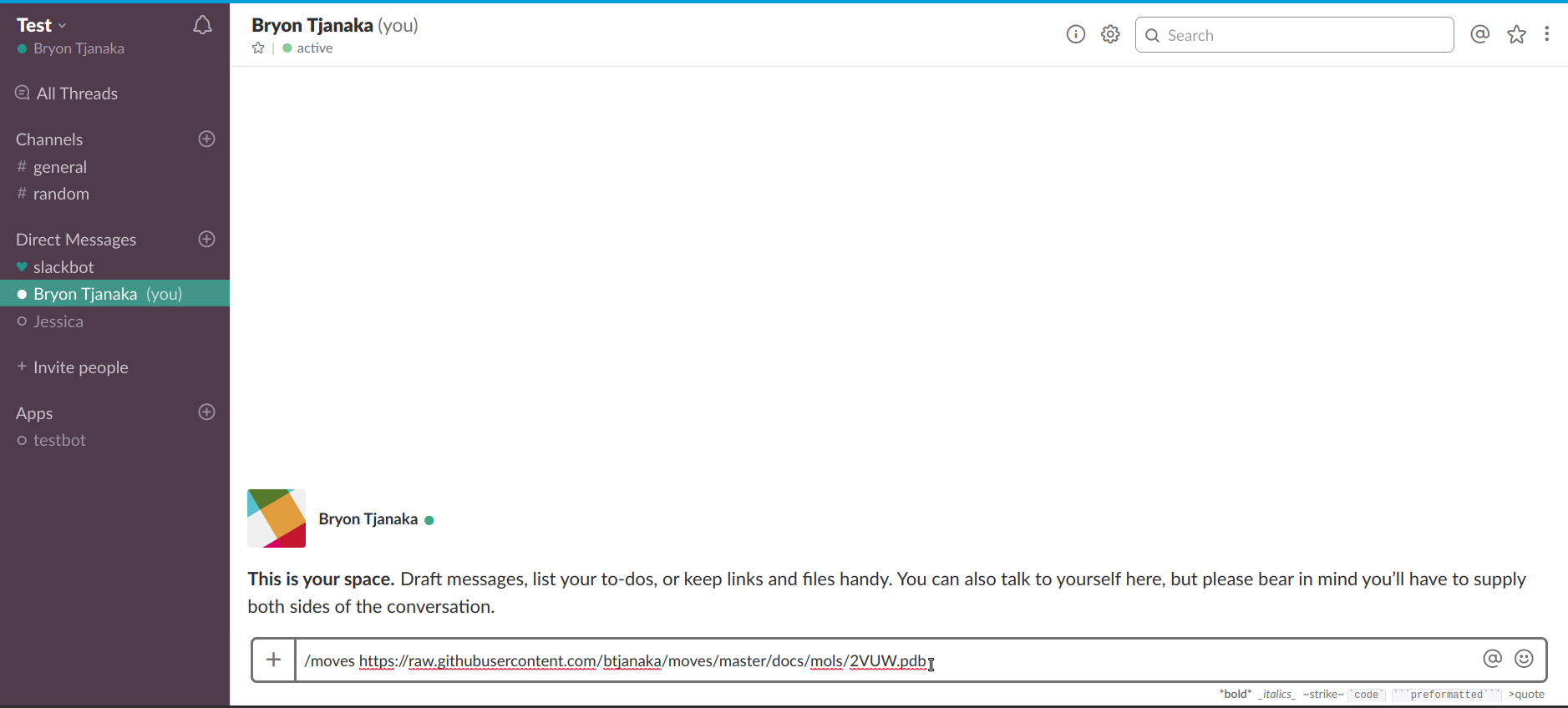
Go to Slack and enter the command "/moves". Then, paste the link and send the message.
You should now have the link!
See CONTRIBUTING.md
This project was completed under the auspices of the Mobley Lab. Many thanks to Dr. Mobley, Jessica Maat, and the entire lab!