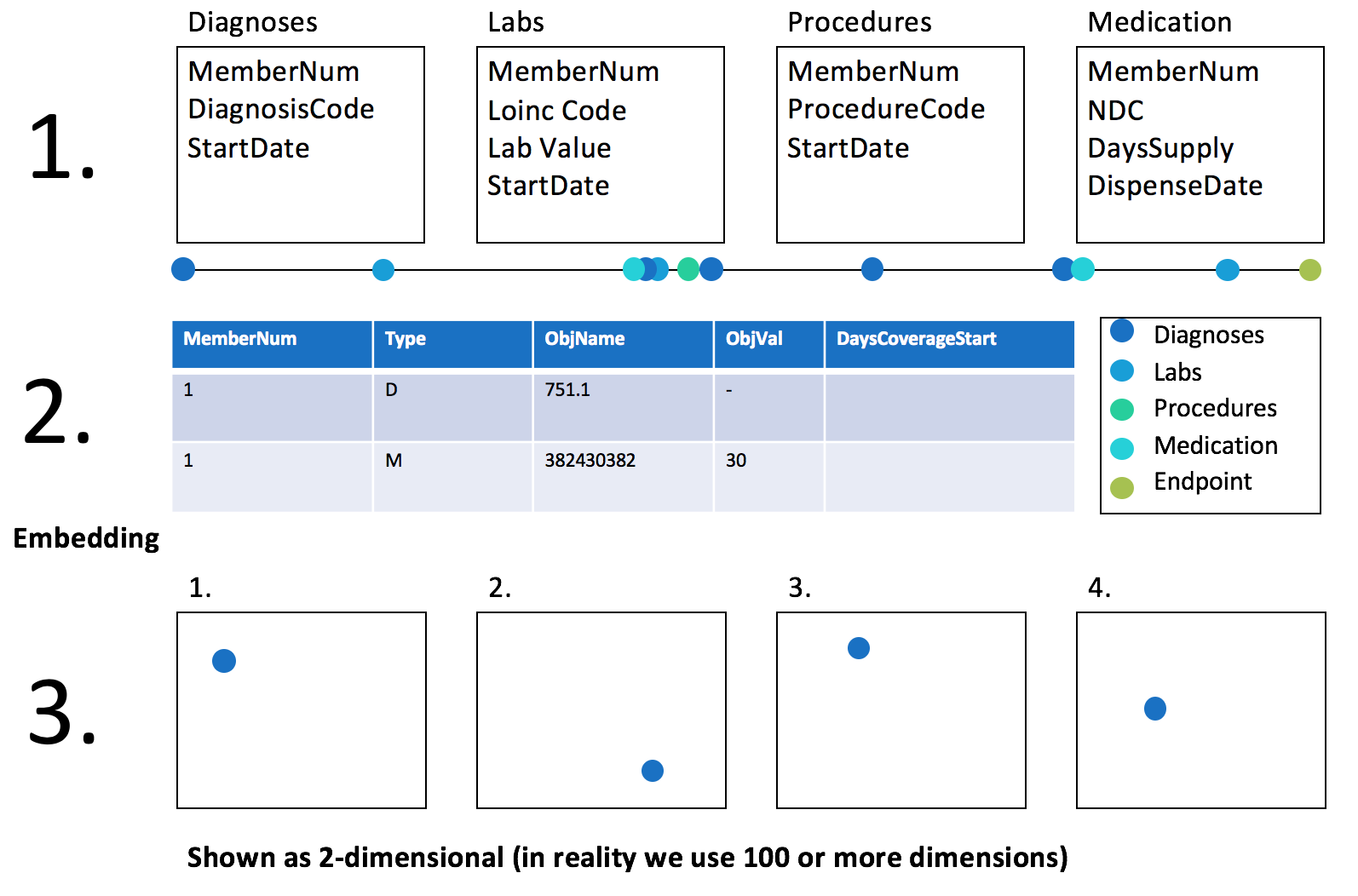
This repository contains code to perform dimensionality reduction of a variable length sequence and then cluster.
Unsupervised clustering is increasingly important for precision medicine and is being used to identify disease subtypes, predict outcomes based on similar patients and identify candidate patients for clinical trials.
Commonly used dimensionality reduction techniques such as Principal Components Analysis, require a fixed length vector and a normally shaped matrix. To fit this, existing methods generally extract fixed vectors at a specific point in time or aggregate vectors using statistics to summarize change over time. Choosing a specific point in time can create a conflict between including a large sample size with low follow-up time or a small sample size with longer follow-up time. Aggregating vectors across different lengths of time can lead to time being a major factor in cluster position.
We introduce a multi-task sequence to sequence deep neural network capable of receiving variable length inputs and correcting for the time since disease onset. We evaluate this network based off of its to produce useful features in a reduced dimension for unsupervised clustering.
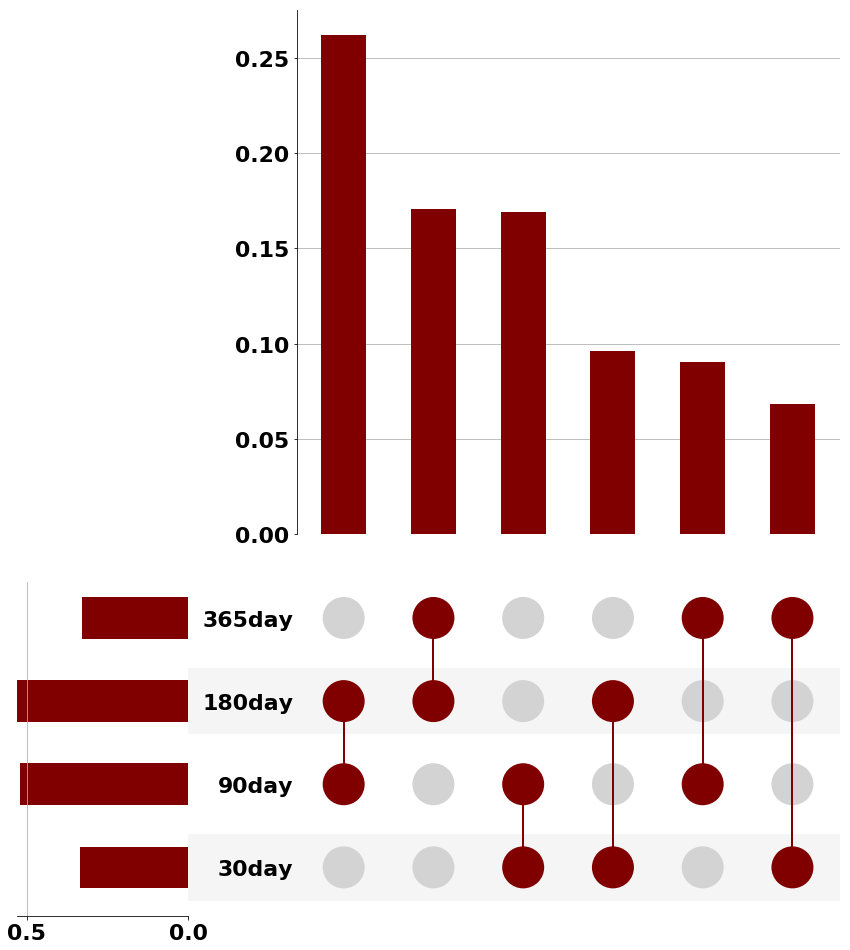
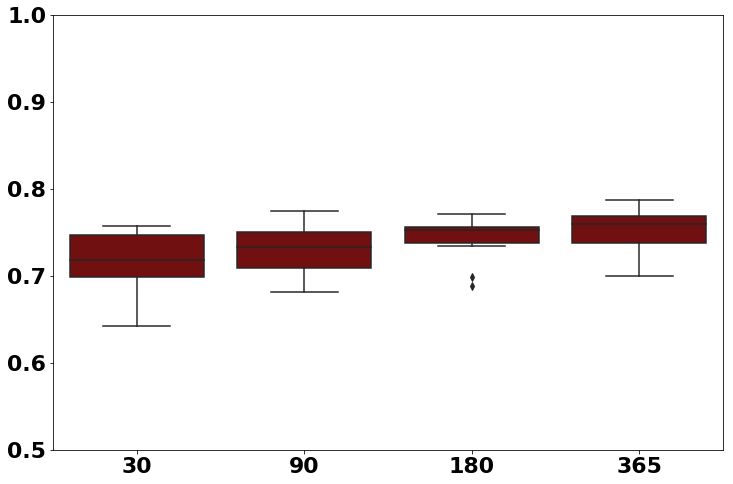
Traditional comorbidity clustering is not stable over time. In this example we show the intersection of neighbors over time (despite having reasonable classification accuracy.)
There are five main parts to this process:
-
Preprocessing - Feature Selection, Construction and learning of distributed representations (embeddings).
- Feature Selection - we currently include 3 settings - comorbidity, fact selection and sequences.
- Construction
- Learning of embeddings - Initially we show typical word2vec embeddings trained with a fixed window.
- Future work includes dynamic bernoulli embeddings, hiearchical representations, context-based representations and
- We perform two separate analyses - include patient histories prior until 0-6 months (uniform) before diagnosis of disease, as well sep , include complete patient histories but exclude related diagnoses from training data.
- For comparison we also build comorbidity matrices as well as fact summarization matrices
-
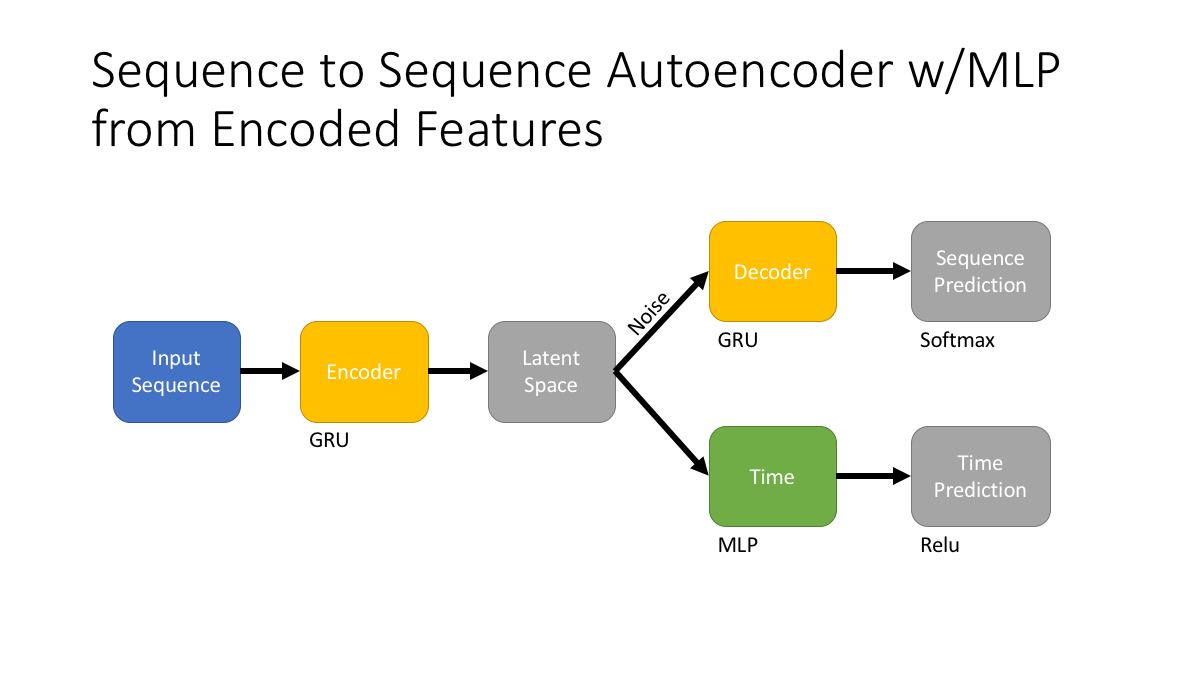
Training the dimensionality reduction (variable to fixed length) model. - Architecture - We pass the input sequences to a seq2seq style model.
-
Performing clustering - Traditional Clustering Techniques:
- KMeans
- Affinity Propagation
- MeanShift
- Spectral Clustering
- Ward
- Agglomerative Clustering
- DBSCAN
- Birch
- Gaussian Mixture Models
- Tested with different Dimensionality Reduction Methods
- Seq2seq dimensionality reduction
- PCA
- UMAP

-
Evaluation - Held-out ICD9 codes - We measure enrichment of held-out labels (ICD9 codes) in identified clusters.
-
Analysis - - Latent space evaluation - we also look for meaningful transformations in the latent space by passing held out sequences in all of their combinations (in attempt to identify the effect of time) as well as passing in each sequence with the opposite gender attached.
We identify all members who had a minimum of one year of coverage before their initial diagnosis for a disease of interest (the index date) and at least two years of coverage after the index date. This quiescence period is designed to filter out those members who have been previously diagnosed for the disease of interest and should be altered based on the diseases of interest. Members are broken into three independent sets, 1.) a training set used to train the Seq2Seq Model, 2.) a validation set used to identify the time vector in the latent space, and 3.) a test set to evaluate the effectiveness of the model.
The initial diagnosis for the ICD code of interest is set as the index date. We then select the 24 concepts (padding for any that do not exist) and placed a neutral event (the same value for all diseases) code in the 25th place of the vector. We calculated the 100 most common diagnoses that occur more than three days after the index date and limit our prediction task to these diseases. The 25 concept vector is the initial input with three different labels, one for each task: 1.) the next five common diagnoses, 2.) the input sequence, 3.) and the time from the index date. This process is repeated for each time step moving forward (moving the index diagnosis into the 24th place and so on) until there are not enough diagnoses to fill the label vector.
- keras 2.1.2
- pandas 0.23.0
- numpy 1.14.5
- scikit-learn 0.19.1
- scipy 1.1.0
- seaborn 0.8.1
- tensorflow-gpu 1.4.1
Coming Soon
Coming soon
Please feel free to email me - (brett_beaulieu-jones) at hms.harvard.edu with any feedback or raise a github issue with any comments or questions.
My work is supported by a T15 NLM Grant (4T15 LM007092-25) and funding from UCB. This repository was built while attending Deep Learning Jeju organized by Tensorflow Korea (sponsored by Google, ).