LocaleDB
A database of global locales to support modeling and simulation in epidemiology with the current focus on the COVID 19 pandemic.
Abstract
Many models of COVID-19 have been proposed to date. When compared against each other, it appears that many of these models make mutually incompatible predictions. Such discrepancies between the output of competing models and simulations need explanation. Inspecting the source code of those models in search for differences which might turn out to be the antecedents of the observed differences is a natural tendency. Scrutinizing source code, however, especially when done automatically, is a valid approach only if model inputs are guaranteed to be identical. For example, if two models use different subsets of the available disease dynamics data, they will likely yield different results even if the epidemiological and social (and other) mechanisms are modeled identically. Moreover, even if the two models are indistinguishable in terms of how they model the various dynamics, comparing them will be hard if they are used to predict the trajectory of an epidemic in distinct locales (e.g., two different U.S. states or counties). The LocaleDB initiative aims to unify, homogenize, and standardize various data sources that are inputs to models of the COVID-19 pandemic and thus provide data control for computational experiments of mass model comparison.
Design
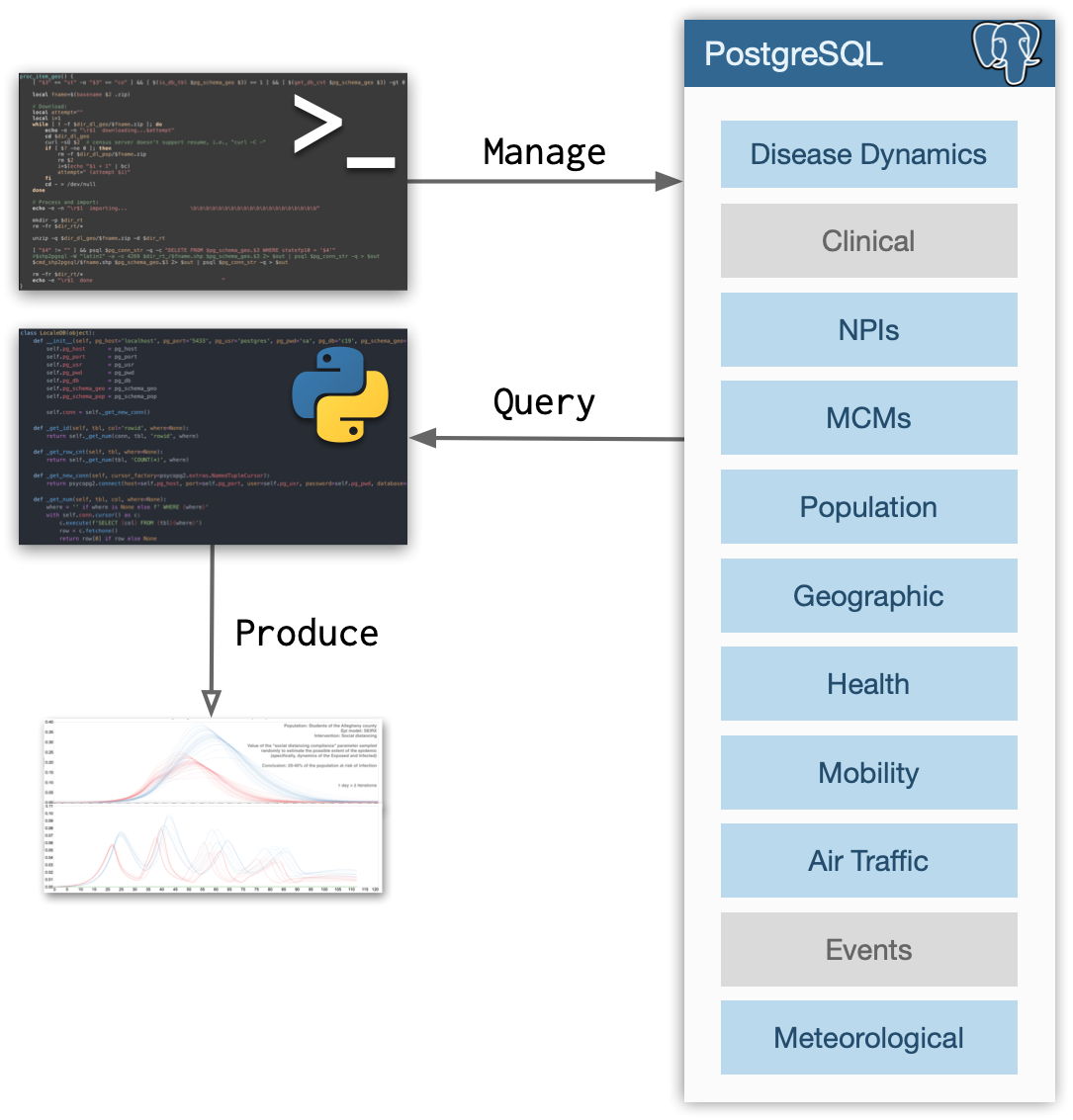
As shown on the figure below, LocaleDB stores several types of data (gray boxes indicate planned future extensions). That data is stored in a PostgreSQL database which is managed by a command line tool (localedb) and a Python script (localedb_man.py). The database can be accessed either via a PostgreSQL driver or a language-specific binding which provide a high level API to, for example, suggest U.S. counties similar to the county specified. To that effect, we are currently working on Python bindings.
This design that separates data management and data consumption reflects the anticipated production use case. Namely, the database will be deployed and set up once and will then require little to no manual management (periodic updates will be autonomous). It will then be used for producing data that will drive modeling and simulation efforts.
Data Types
As depicted on the figure above, the current projection is for LocaleDB to contain the following data types:
- Disease dynamics (e.g., number of confirmed cases)
- Clinical (e.g., R0, incubation period, proportion of asymptomatic cases, etc.)
- Non-pharmaceutical interventions (NPIs; e.g., dates of stay-at-home order)
- Medical countermeasures (MCMs; e.g., vaccine availability, efficacy, and allocation strategies)
- Population (e.g., households, their incomes, age of people, etc.)
- Geographic and cartographic (e.g., area of land, population density)
- Health factors and outcomes (e.g., diet, exercise, access to care, etc.)
- Mobility (e.g., number of trips taken by individuals based on mobile-phone data)
- Air Traffic (e.g., number of passengers landing in US cities; includes origin and destination)
- Local events (e.g., dates and sizes of mass protests)
- Meteorological (e.g., monthly temperature and precipitation)
All that data is stratified by locale at all available levels of spatial aggregation (e.g., country, state, county, tract, block group, block). In terms of temporal resolution, the highest frequency with which processes are sampled/measured is the goal. For example, disease dynamics is represented as a time series of daily numbers of confirmed cases and deaths, while health factors and outcomes are encoded with far fewer time steps.
Dependencies
Development/Test Environment
- curl or wget
- Docker
Production Environment
- curl or wget
- PostgreSQL client
- PostgreSQL server (with PostGIS and TimescaleDB extensions)
- Python 3
Note: Full production environment support will be added at a later date.
Setup and Example Usage: Development/Test Environment
The development environment is based around Docker so you will need to get that installed first. After sorting that out, clone the repo and change the working directory. The cloned repo directory will be where all the data is stored so if you plan on loading a whole lot (looking at you, geo and pop data users), make sure you have enough space on that filesystem subtree.
git clone https://github.com/momacs/localedb
cd localedb
Then:
# Build the container:
./build-docker.sh
# Start the container (takes about a minute for Postgres to become available):
docker-compose up -d
# Initialize the database inside the container:
docker-compose run --rm localedb setup
Displaying info and loading data is done using the following commands:
# Display info:
docker-compose run --rm localedb info all
# Load disease data for COVID-19:
docker-compose run --rm localedb load dis COVID-19
# Load Alaska geographic and cartographic data:
docker-compose run --rm localedb load geo AK
# Load synthetic population for Alaska:
docker-compose run --rm localedb load pop AK
# Load health data for Alaska:
docker-compose run --rm localedb load health AK
# Load weather data for years 2010 through 2020:
docker-compose run --rm localedb load weather 2010 2020
# Load vaccination data:
docker-compose run --rm localedb load vax
# Load mobility data for Alaska:
docker-compose run --rm localedb load mobility AK
# Load air traffic data for 2019 for flights traveling to Alaska (only flights with 25+ passengers):
docker-compose run --rm localedb load airtraffic 2019 AK 25
For the list of available commands, run either of the two:
docker-compose run --rm localedb
docker-compose run --rm localedb help
Keep in mind that some commands have subcommands.
Once not needed any more, the container can be stopped or taken down (i.e., stopped, removed, and all networks created removed):
docker-compose stop --timeout 300
docker-compose down --timeout 300
To remove the container with all associated volumes (watch out with this one because this includes the data you have loaded!), do:
docker-compose down -v --timeout 300
Grafana
Running LocaleDB with Docker (i.e., the development environment) has the added benefit of automatically deploying a Grafana instance. Grafana provides an open source data visualization and dashboarding platform to view and analyze LocaleDB. By default, it runs at http://localhost:3000 so head there once you get the Docker container up.
Currently, only the vaccination data dashboard is provided as an example. Naturally, to be able to view any data on that dashboard, you'll need to load the data first:
docker-compose run --rm localedb load vax
To create a new dashboard, do so via the Grafana UI and export it as a JSON file. Save this JSON file to grafana/dashboards.
Development with Docker
Any changes to the docker image require ./build-docker.sh to be run to update the image. localedb and localedb_man.py are mounted as volumes, so any changes to the management and loading routines will take effect immediately.
Setup and Example Usage: Production Environment
Command Line Management Tool
On MacOS run:
sh -c "$(curl -fsSL https://raw.githubusercontent.com/momacs/localedb/master/setup.sh -O -)"
On Linux run:
sh -c "$(wget -q https://raw.githubusercontent.com/momacs/localedb/master/setup.sh -O -)"
Alternatively, you can run the commands from the setup.sh script manually.
Production environment: For production deployment, after the installation script above has finished, edit the $HOME/bin/localedb script and change is_prod=0 to is_prod=1. This step is left to be done manually to ensure intent.
CLI
After setting up the command line management tool, setup the LocaleDB instance:
$ localedb setup
Initializing data structures... done
Loading locales... done
To display filesystem information, run:
$ localedb info fs
Directory structure
Root /Users/tomek/.localedb 43M
Runtime /Users/tomek/.localedb/rt 0B
PostgreSQL data /Users/tomek/.localedb/pg 43M
Disease data /Users/tomek/.localedb/dl/dis 0B
Geographic data /Users/tomek/.localedb/dl/geo 0B
Population data /Users/tomek/.localedb/dl/pop 0B
To import COVID-19 disease data (currently only dynamics and non-pharmaceutical interventions), run:
$ localedb import dis c19
Disease dynamics
Loading global confirmed... done (11 s)
Loading global deaths... done (14 s)
Loading global recovered... done (12 s)
Loading US confirmed... done (141 s)
Loading US deaths... done (155 s)
Consolidating... done (88 s)
Non-pharmaceutical interventions
Loading Keystone... done (14 s)
To see some basic database statistics, run:
$ localedb info data
Data
Main
Locale count 4153
Country count 188
Disease (c19)
Dynamics
Locale count 3607
Date range 2020-01-22 2020-09-17
Observation count 865680
Observation count per locale 240.00 (SD=0.00)
Non-pharmaceutical interventions
Locale count 669
Data range 2010-04-27 2020-07-27
NPI count 5162
NPI count per locale 7.72 (SD=1.57)
Count per type
669 school closure
667 closing of public venues
666 non-essential services closure
637 shelter in place
622 gathering size 10 0
582 social distancing
471 religious gatherings banned
406 gathering size 100 26
278 gathering size 500 101
132 gathering size 25 11
32 lockdown
...
To import geographic and cartographic data for the state of Alaska, run:
$ localedb load geo AK
US states done
US counties done
AK tracts done
AK block groups done
AK blocks done
Analyzing database... done
To import health data for the state of Alaska, run:
$ localedb load health AK
To import weather data from 2010 to 2020, run:
$ localedb load weather 2010 2020
To import mobility data for Alaska run:
$ localedb load mobility AK
To import air traffic data for 2019 for flights travelling to Alaska which had more than 25 passengers run:
$ localedb load airtraffic 2019 AK 25
To import synthetic population data, run:
$ localedb load pop AK
AK done
Analyzing database... done
Imported states can be removed like so:
localedb db rm state-geo AK
localedb db rm state-pop AK
localedb db rm state AK # remove all data types
Once data has been imported and the downloaded data files are no longer needed, they can be removed like so:
localedb fs rm-data geo
localedb db rm-data pop
localedb db rm data-all # remove all data files
To stop LocaleDB instance, run:
localedb stop
To uninstall LocaleDB (leaving nothing behind), run:
localedb uninstall
For the list of available commands, run localedb. For an explanation of each command, run localedb help. Keep in mind that some commands have subcommands.
References
Data Sources
- COVID-19 Data Repository by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University
- CDC: Influenza Vaccination Coverage
- County Health Rankings and Roadmaps
- Bureau of Transportation Statistics: Airlines and Airports
- NOAA nClimDiv
- Keystone: COVID-19 Intervention Data
- 2010 U.S. Synthesized Population Dataset
- US Census Bureau: TIGER/Line Shapefiles (2010)
License
This project is licensed under the BSD License.