This project is for Deep Neighbor Layer Aggregation for Lightweight Self-Supervised Monocular Depth Estimation ICASSP 2024
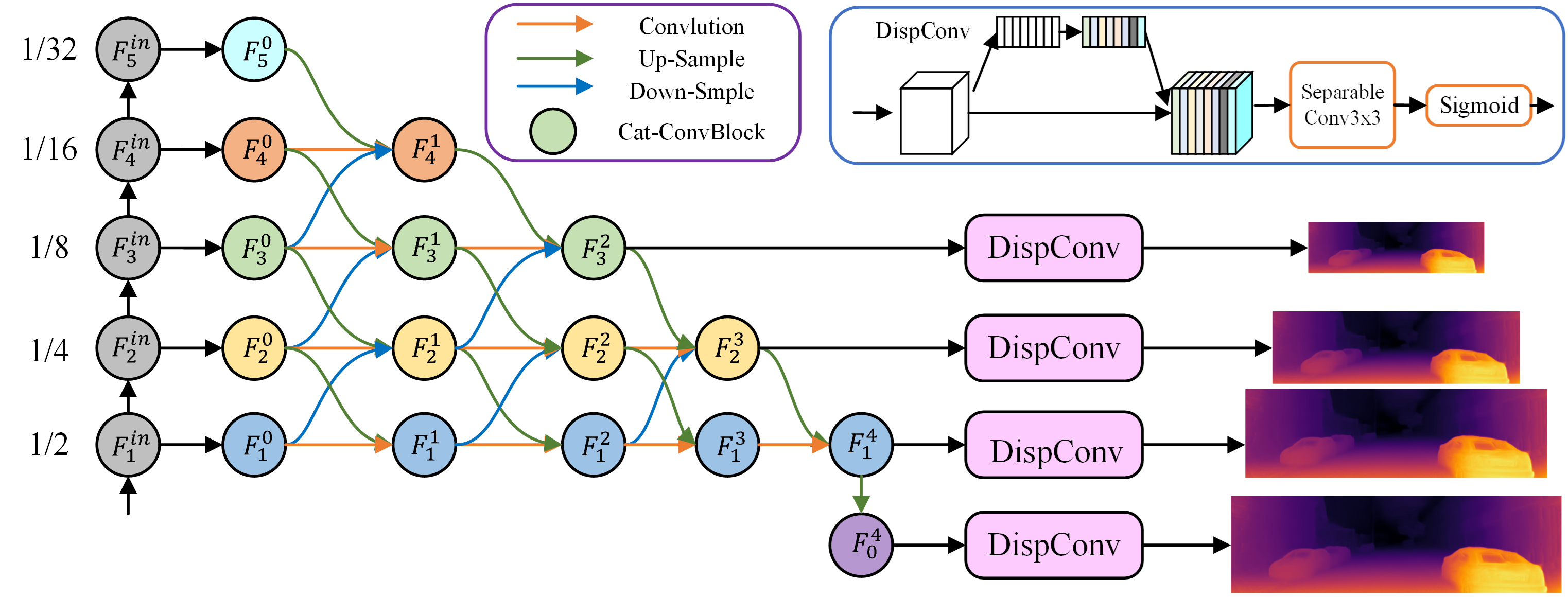
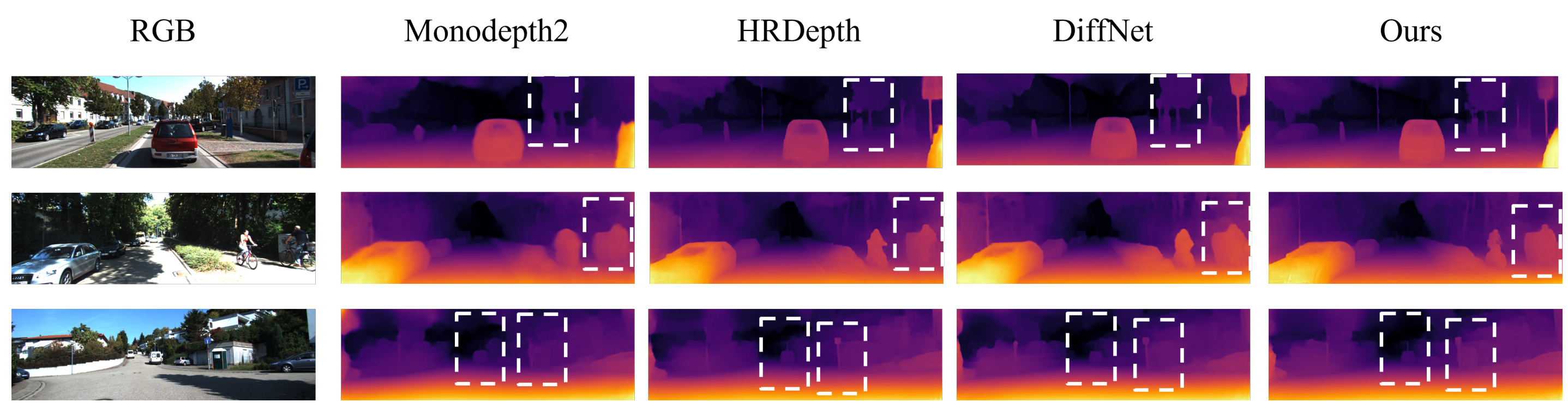
This code uses PyTorch implementation for training and testing depth estimation models. The schematic of the network is shown below:
Assuming a fresh Anaconda distribution, you can install the dependencies with:
conda create -n dna python=3.6
conda activate dna
conda install six matplotlib scikit-image ipython
conda install pytorch==1.7.0 torchvision==0.8.0 torchaudio==0.7.0 cudatoolkit=11.0 -c pytorch
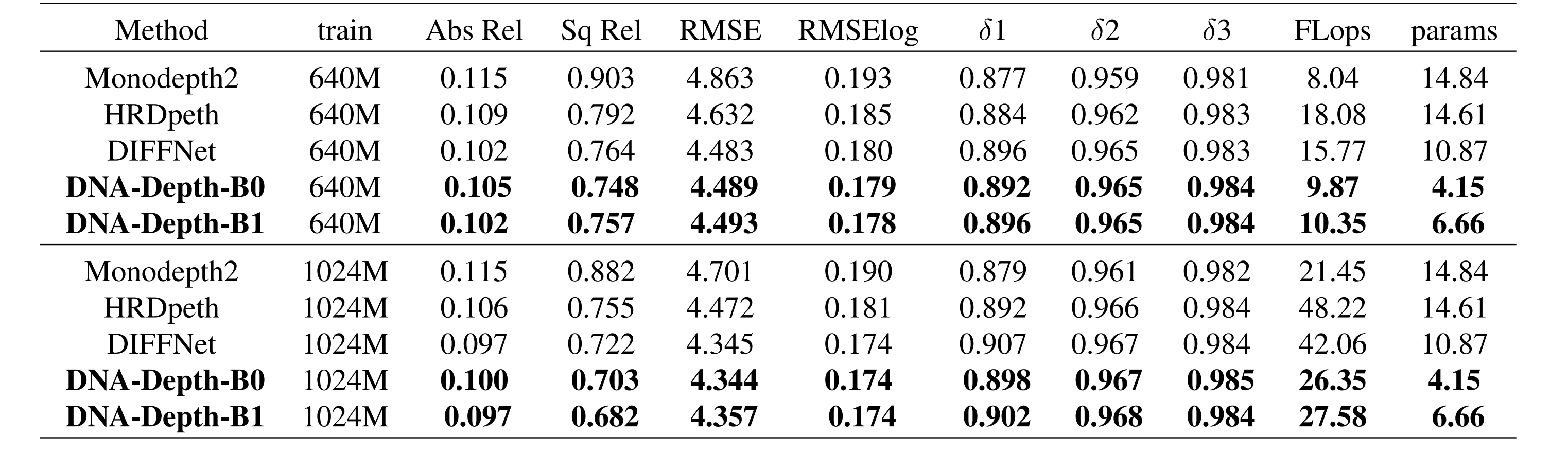
pip install tensorboardX==2.2 opencv-python==4.5.3.56python train.py python evaluate_depth.py --load_weights_folder mono_640m_b0 --eval_mono --png
--model_name |
Model resolution | KITTI abs. rel. error | delta < 1.25 | FLops | params |
|---|---|---|---|---|---|
mono_640m_b0 |
640 x 192 | 0.105 | 0.892 | 9.87 | 4.15 |
mono_640m_b1 |
640 x 192 | 0.102 | 0.896 | 10.35 | 6.66 |
mono_1024m_b0 |
1024 x 320 | 0.100 | 0.898 | 26.35 | 4.15 |
mono_1024m_b1 |
1024 x 320 | 0.097 | 0.902 | 27.58 | 6.66 |
Thanks the authors for their works: