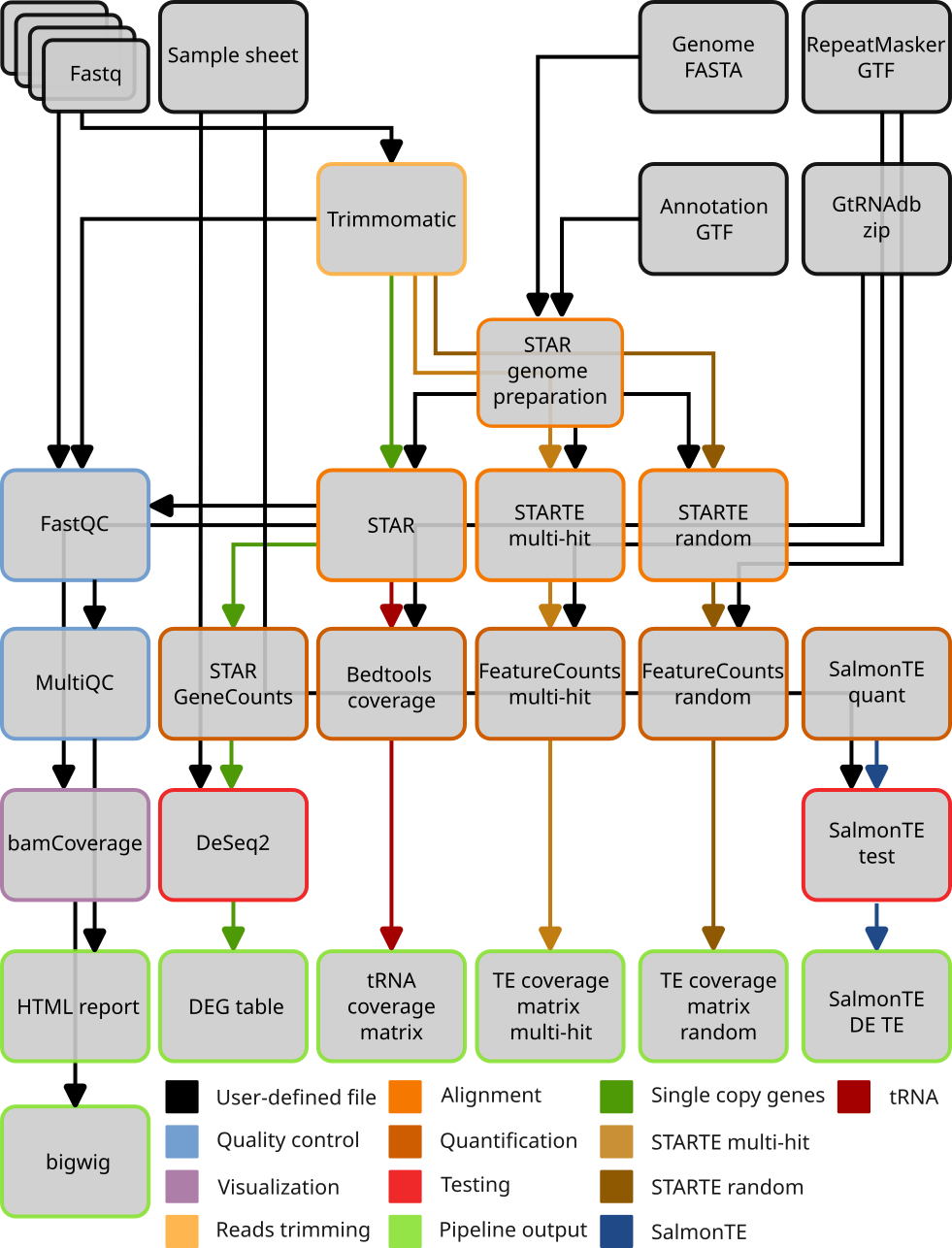
This is a Snakemake workflow for the integrated analysis of single copy genes, transposable elements and tRNAs. It performs standard quality control checks and genome alignment in three different ways specialized either for single copy genes or transposable elements. It then quantifies gene expression depending on how the alignement step was performed. Finally it performs differential gene expression analysis yielding lists of genes significantly deregulated between two given conditions.
For example, to download version 1.0.0:
curl -LJO "https://github.com/boulardlab/3t-seq/archive/v1.0.0.zip"
unzip "3t-seq-1.0.0.zip"conda create -n snakemake-latest -c bioconda -c conda-forge snakemake singularity
conda activate snakemake-latestEdit the config.yaml file to specify your sample information and analysis parameters. The config/ folder contains a detailed description of this file.
In the 3et-seq folder:
snakemake --profile profile/defaultAfter the pipeline completes, you can find the results in the results/ directory.
After successful execution, an interactive HTML report collecting execution statistics, FastQC and MultiQC reports, DESeq2 results, MA plot and Volcano plots for single copy gene, retrotransposons and tRNAs can be generated as follow:
snakemake --profile profile/default --report report.zipA report.zip archive will be generated in the current working directory. The archive will contain the HTML file. This file can be shared and does not need internet connection to be opened.
Adjust parameters in the config.yaml file to match your experimental setup. See config/README.md for further instructions.
The sample sheet is a csv file that describe samples metadata:
- The
samplecolumn reports a human readable name for each sample. - For pe libraries,
filename_1andfilename_2columns report file names for each of the two sequencing reads mates. For se libraries,filenameis sufficient. The pipeline will use these columns to determine if a given dataset was sequenced with pe or se method. - The
genotypecolumn reports the variable of interest. The name of this column is flexible and can be anything as long as you specify what's this name in the config file (in thedeseq2section).
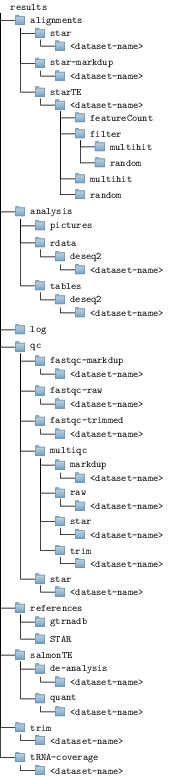
The pipeline will generate an ouput folder tree like so
The tests/ folder contains a small test dataset and example configuration file needed to run the 3t-seq pipeline on it.
Provided a working Snakemake installation is available, the example dataset can be run as follow:
snakemake \
--directory tests \
--configfile tests/config.yaml \
--profile tests/profile \
--snakefile workflow/SnakefileResults will then be available in tests/results.
An example 3t-seq HTML report could be generated with the following command:
snakemake \
--directory tests \
--configfile tests/config.yaml \
--profile tests/profile \
--snakefile workflow/Snakefile \
--report report.zipThe report.zip file will be generated in tests/report.zip.
Tabaro F, Boulard M, 3t-seq: automatic gene expression analysis of single copy genes, transposable elements and tRNAs from total RNA-seq data, Under review.
This project is licensed under the MIT License.