This package contains a marginal likelihood approach to estimating the model discussed by Hoff (2007), Zhao and Udell (2020b), and Zhao and Udell (2020a). That is, a missing data approach where one uses Gaussian copulas in the latter case. We have modified the Fortran code by Genz and Bretz (2002) to supply an approximation of the gradient for the log marginal likelihood and to use an approximation of the marginal likelihood similar to the CDF approximation in Genz and Bretz (2002). We have also used the same Fortran code to perform the imputation conditional on a covariance matrix and the observed data. The method is described by Christoffersen et al. (2021) which can be found at arxiv.org.
Importantly, we also extend the model used by Zhao and Udell (2020b) to support multinomial variables. Thus, our model supports both continuous, binary, ordinal, and multinomial variables which makes it applicable to a large number of data sets.
The package can be useful for a lot of other models. For instance, the methods are directly applicable to other Gaussian copula models and some mixed effect models. All methods are implemented in C++, support computation in parallel, and should easily be able to be ported to other languages.
The package can be installed from Github by calling:
remotes::install_github("boennecd/mdgc")or from CRAN by calling:
install.packages("mdgc")The code benefits from being build with automatic vectorization so
having e.g.
-O3 -mtune=native in the CXX11FLAGS flags in your Makevars file may
be useful.
We observe four types of variables for each observation: continuous,
binary, ordinal, and multinomial variables. Let be a K dimensional vector for the i’th observation. The
variables
are continuous if
, binary if
with probability
of being true, ordinal if
with
levels and borders
,
and multinomial if
with
levels.
,
,
, and
are mutually exclusive.
We assume that there is a latent variable which is multivariate normally distributed such that:
where
is one if the condition in the subscript is true and
zero otherwise,
is a map to the index of the first latent variable associated
with the j’th variable in
and
is a bijective function. We only estimate some of the means, the
, and some of the covariance parameters. Furthermore, we set
if
and assume that the variable is uncorrelated with
all the other
’s.
In principle, we could use other distributions than a multivariate
normal distribution for . However, the multivariate normal distribution has the
advantage that it is very easy to marginalize which is convenient when
we have to estimate the model with missing entries and it is also has
some computational advantages for approximating the log marginal
likelihood as similar intractable problem have been thoroughly studied.
Below, we provide an example similar to Zhao and Udell (2020b Section 7.1). The authors use a data set with a random correlation matrix, 5 continuous variables, 5 binary variables, and 5 ordinal variables with 5 levels. There is a total of 2000 observations and 30% of the variables are missing completely at random.
To summarize Zhao and Udell (2020b) results, they show that their approximate EM algorithm converges in what seems to be 20-25 seconds (this is with a pure R implementation to be fair) while it takes more than 150 seconds for the MCMC algorithm used by Hoff (2007). These figures should be kept in mind when looking at the results below. Importantly, Zhao and Udell (2020b) use an approximation in the E-step of an EM algorithm which is fast but might be crude in some settings. Using a potentially arbitrarily precise approximation of the log marginal likelihood is useful if this can be done quickly enough.
We will provide a quick example and an even shorter example where we show how to use the methods in the package to estimate the correlation matrix and to perform the imputation. We then show a simulation study where we compare with the method suggested by Zhao and Udell (2020b).
The last section called adding multinomial variables covers data sets which also have multinomial variables.
We first simulate a data set and provide an example which shows how to use the package. The an even shorter example section shows a shorter example then what is shown here. You may want to see this first if you just want to perform some quick imputation.
# load the packages we need
library(bench)
library(mdgc)
library(missForest, quietly = TRUE)
#> randomForest 4.6-14
#> Type rfNews() to see new features/changes/bug fixes.
# remotes::install_github("udellgroup/mixedgcImp", ref = "5ad6d523d")
library(mixedgcImp)
library(doParallel)
#> Loading required package: parallel# simulates a data set and mask some of the data.
#
# Args:
# n: number of observations.
# p: number of variables.
# n_lvls: number of levels for the ordinal variables.
#
# Returns:
# Simulated masked data, the true data, and true covariance matrix.
sim_dat <- function(n, p = 3L, n_lvls = 5L){
# get the covariance matrix
Sig <- cov2cor(drop(rWishart(1L, p, diag(p))))
# draw the observations
truth <- matrix(rnorm(n * p), n) %*% chol(Sig)
# determine the type
n_rep <- floor((p + 3 - 1) / 3)
type <- rep(1:3, each = n_rep)[1:p]
is_con <- type == 1L
is_bin <- type == 2L
is_ord <- type == 3L
col_nam <- c(outer(1:n_rep, c("C", "B", "O"),
function(x, y) paste0(y, x)))[1:p]
# sample which are masked data
is_mask <- matrix(runif(n * p) < .3, n)
# make sure we have no rows with all missing data
while(any(all_nans <- rowSums(is_mask) == NCOL(is_mask)))
is_mask[all_nans, ] <- runif(sum(all_nans) * p) < .3
# create observed data
truth_obs <- data.frame(truth)
colnames(truth_obs) <- col_nam
truth_obs[, is_con] <- qexp(pnorm(as.matrix(truth_obs[, is_con])))
bs_border <- 0
truth_obs[, is_bin] <-
truth_obs[, is_bin] > rep(bs_border, each = NROW(truth_obs))
bs_ord <- qnorm(seq(0, 1, length.out = n_lvls + 1L))
truth_obs[, is_ord] <- as.integer(cut(truth[, is_ord], breaks = bs_ord))
for(i in which(is_ord)){
truth_obs[, i] <- ordered(truth_obs[, i])
levels(truth_obs[, i]) <-
LETTERS[seq_len(length(unique(truth_obs[, i])))]
}
# mask the data
seen_obs <- truth_obs
seen_obs[is_mask] <- NA
list(truth = truth, truth_obs = truth_obs, seen_obs = seen_obs,
Sigma = Sig)
}
# simulate and show the data
set.seed(1)
p <- 15L
dat <- sim_dat(2000L, p = p)
# how an observed data set could look
head(dat$seen_obs)
#> C1 C2 C3 C4 C5 B1 B2 B3 B4 B5 O1 O2
#> 1 0.237 0.693 0.798 0.0666 NA FALSE FALSE FALSE FALSE TRUE E C
#> 2 0.142 NA NA 0.0927 0.000152 FALSE NA TRUE NA NA E B
#> 3 NA 0.748 0.629 0.4280 NA NA TRUE NA NA TRUE <NA> A
#> 4 2.702 NA NA 2.1776 1.700870 FALSE TRUE TRUE NA TRUE A <NA>
#> 5 0.925 NA 0.205 0.6046 0.171311 TRUE TRUE FALSE FALSE FALSE E B
#> 6 0.115 NA 1.341 NA NA FALSE TRUE TRUE NA NA E <NA>
#> O3 O4 O5
#> 1 B B <NA>
#> 2 A A C
#> 3 <NA> C E
#> 4 D B <NA>
#> 5 <NA> D <NA>
#> 6 A B <NA>
# assign objects needed for model estimation
mdgc_obj <- get_mdgc(dat$seen_obs)
log_ml_ptr <- get_mdgc_log_ml(mdgc_obj)
start_val <- mdgc_start_value(mdgc_obj)
# this is very fast so we can neglect this when we consider the computation
# time
mark(`Setup time` = {
mdgc_obj <- get_mdgc(dat$seen_obs)
log_ml_ptr <- get_mdgc_log_ml(mdgc_obj)
start_val <- mdgc_start_value(mdgc_obj)
}, min_iterations = 10)
#> # A tibble: 1 x 6
#> expression min median `itr/sec` mem_alloc `gc/sec`
#> <bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>
#> 1 Setup time 16.4ms 17.1ms 57.7 9.57MB 23.1
# fit the model using three different methods
set.seed(60941821)
system.time(
fit_Lagran_start <- mdgc_fit(
ptr = log_ml_ptr, vcov = start_val, mea = mdgc_obj$means,
n_threads = 4L, maxit = 100L, method = "aug_Lagran", rel_eps = 1e-3,
maxpts = 200L))
#> user system elapsed
#> 134.24 0.02 33.58
system.time(
fit_Lagran <- mdgc_fit(
ptr = log_ml_ptr, vcov = fit_Lagran_start$result$vcov,
mea = fit_Lagran_start$result$mea,
n_threads = 4L, maxit = 100L, method = "aug_Lagran", rel_eps = 1e-3,
maxpts = 5000L, mu = fit_Lagran_start$mu,
lambda = fit_Lagran_start$lambda))
#> user system elapsed
#> 31.887 0.016 8.113
system.time(
fit_adam <- mdgc_fit(
ptr = log_ml_ptr, vcov = start_val, mea = mdgc_obj$means,
n_threads = 4L, lr = 1e-3, maxit = 25L, batch_size = 100L,
method = "adam", rel_eps = 1e-3, maxpts = 5000L))
#> user system elapsed
#> 30.786 0.008 7.722
set.seed(fit_seed <- 19570958L)
system.time(
fit_svrg <- mdgc_fit(
ptr = log_ml_ptr, vcov = start_val, mea = mdgc_obj$means,
n_threads = 4L, lr = 1e-3, maxit = 25L, batch_size = 100L,
method = "svrg", verbose = TRUE, rel_eps = 1e-3, maxpts = 5000L))
#> End of iteration 1 with learning rate 0.00100000
#> Log marginal likelihood approximation is -23439.04
#> Previous approximate gradient norm was 3446.50
#>
#> End of iteration 2 with learning rate 0.00098000
#> Log marginal likelihood approximation is -23388.66
#> Previous approximate gradient norm was 1736.96
#>
#> End of iteration 3 with learning rate 0.00096040
#> Log marginal likelihood approximation is -23366.80
#> Previous approximate gradient norm was 1149.42
#>
#> End of iteration 4 with learning rate 0.00094119
#> Log marginal likelihood approximation is -23355.07
#> Previous approximate gradient norm was 846.52
#>
#> End of iteration 5 with learning rate 0.00092237
#> Log marginal likelihood approximation is -23348.13
#> Previous approximate gradient norm was 665.70
#>
#> End of iteration 6 with learning rate 0.00090392
#> Log marginal likelihood approximation is -23343.76
#> Previous approximate gradient norm was 547.47
#>
#> End of iteration 7 with learning rate 0.00088584
#> Log marginal likelihood approximation is -23340.79
#> Previous approximate gradient norm was 468.29
#>
#> End of iteration 8 with learning rate 0.00086813
#> Log marginal likelihood approximation is -23338.65
#> Previous approximate gradient norm was 411.70
#>
#> End of iteration 9 with learning rate 0.00085076
#> Log marginal likelihood approximation is -23337.10
#> Previous approximate gradient norm was 370.40
#>
#> End of iteration 10 with learning rate 0.00083375
#> Log marginal likelihood approximation is -23335.93
#> Previous approximate gradient norm was 337.65
#>
#> End of iteration 11 with learning rate 0.00081707
#> Log marginal likelihood approximation is -23335.04
#> Previous approximate gradient norm was 315.33
#>
#> End of iteration 12 with learning rate 0.00080073
#> Log marginal likelihood approximation is -23334.31
#> Previous approximate gradient norm was 297.13
#>
#> End of iteration 13 with learning rate 0.00078472
#> Log marginal likelihood approximation is -23333.74
#> Previous approximate gradient norm was 282.12
#>
#> End of iteration 14 with learning rate 0.00076902
#> Log marginal likelihood approximation is -23333.26
#> Previous approximate gradient norm was 268.49
#>
#> End of iteration 15 with learning rate 0.00075364
#> Log marginal likelihood approximation is -23332.87
#> Previous approximate gradient norm was 257.65
#>
#> End of iteration 16 with learning rate 0.00073857
#> Log marginal likelihood approximation is -23332.52
#> Previous approximate gradient norm was 248.55
#>
#> End of iteration 17 with learning rate 0.00072380
#> Log marginal likelihood approximation is -23332.12
#> Previous approximate gradient norm was 281.80
#>
#> End of iteration 18 with learning rate 0.00070932
#> Log marginal likelihood approximation is -23331.85
#> Previous approximate gradient norm was 256.66
#>
#> End of iteration 19 with learning rate 0.00069514
#> Log marginal likelihood approximation is -23331.63
#> Previous approximate gradient norm was 248.70
#>
#> End of iteration 20 with learning rate 0.00068123
#> Log marginal likelihood approximation is -23331.44
#> Previous approximate gradient norm was 243.76
#>
#> End of iteration 21 with learning rate 0.00066761
#> Log marginal likelihood approximation is -23331.33
#> Previous approximate gradient norm was 250.50
#>
#> End of iteration 22 with learning rate 0.00065426
#> Log marginal likelihood approximation is -23331.19
#> Previous approximate gradient norm was 236.98
#>
#> End of iteration 23 with learning rate 0.00064117
#> Log marginal likelihood approximation is -23331.07
#> Previous approximate gradient norm was 232.90
#>
#> End of iteration 24 with learning rate 0.00062835
#> Log marginal likelihood approximation is -23331.02
#> Previous approximate gradient norm was 233.03
#> user system elapsed
#> 54.85 0.02 13.75
# compare the log marginal likelihood
print(rbind(
`Augmented Lagrangian` =
mdgc_log_ml(vcov = fit_Lagran$result$vcov, mea = fit_Lagran$result$mea,
ptr = log_ml_ptr, rel_eps = 1e-3),
ADAM =
mdgc_log_ml(vcov = fit_adam$result$vcov , mea = fit_adam$result$mea,
ptr = log_ml_ptr, rel_eps = 1e-3),
SVRG =
mdgc_log_ml(vcov = fit_svrg$result$vcov , mea = fit_svrg$result$mea,
ptr = log_ml_ptr, rel_eps = 1e-3),
Truth =
mdgc_log_ml(vcov = dat$Sigma , mea = numeric(5),
ptr = log_ml_ptr, rel_eps = 1e-3)),
digits = 10)
#> [,1]
#> Augmented Lagrangian -23330.35309
#> ADAM -23348.70873
#> SVRG -23330.86030
#> Truth -23384.11648
# we can use an approximation in the method
set.seed(fit_seed)
system.time(
fit_svrg_aprx <- mdgc_fit(
ptr = log_ml_ptr, vcov = start_val, mea = mdgc_obj$means,
n_threads = 4L, lr = 1e-3, maxit = 25L, batch_size = 100L,
method = "svrg", rel_eps = 1e-3, maxpts = 5000L, use_aprx = TRUE))
#> user system elapsed
#> 28.18 0.00 7.05
# essentially the same estimates
norm(fit_svrg_aprx$result$vcov - fit_svrg$result$vcov, "F")
#> [1] 8.98e-08
sd(fit_svrg_aprx$result$mea - fit_svrg$result$mea)
#> [1] 2.17e-09
# compare the estimated correlation matrix with the true value
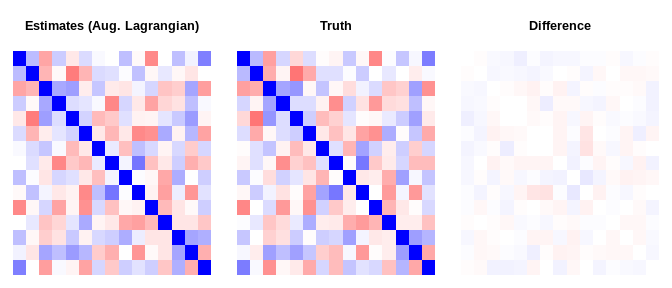
do_plot <- function(est, truth, main){
par_old <- par(mfcol = c(1, 3), mar = c(1, 1, 4, 1))
on.exit(par(par_old))
sc <- colorRampPalette(c("Red", "White", "Blue"))(201)
f <- function(x, main)
image(x[, NCOL(x):1], main = main, col = sc, zlim = c(-1, 1),
xaxt = "n", yaxt = "n", bty = "n")
f(est, main)
f(truth, "Truth")
f(est - truth, "Difference")
}
do_plot(fit_Lagran$result$vcov, dat$Sigma, "Estimates (Aug. Lagrangian)")do_plot(fit_adam $result$vcov, dat$Sigma, "Estimates (ADAM)")do_plot(fit_svrg $result$vcov, dat$Sigma, "Estimates (SVRG)")norm(fit_Lagran$result$vcov - dat$Sigma, "F")
#> [1] 0.486
norm(fit_adam $result$vcov - dat$Sigma, "F")
#> [1] 0.504
norm(fit_svrg $result$vcov - dat$Sigma, "F")
#> [1] 0.488
# perform the imputation
system.time(imp_res <- mdgc_impute(
mdgc_obj, fit_svrg$result$vcov, mea = fit_svrg$result$mea, rel_eps = 1e-3,
maxit = 10000L, n_threads = 4L))
#> user system elapsed
#> 14.19 0.00 3.97
# look at the result for one of the observations
imp_res[2L]
#> [[1]]
#> [[1]]$C1
#> [1] 0.142
#>
#> [[1]]$C2
#> [1] 2.08
#>
#> [[1]]$C3
#> [1] 0.249
#>
#> [[1]]$C4
#> [1] 0.0927
#>
#> [[1]]$C5
#> [1] 0.000152
#>
#> [[1]]$B1
#> FALSE TRUE
#> 1 0
#>
#> [[1]]$B2
#> FALSE TRUE
#> 0.192 0.808
#>
#> [[1]]$B3
#> FALSE TRUE
#> 0 1
#>
#> [[1]]$B4
#> FALSE TRUE
#> 0.81 0.19
#>
#> [[1]]$B5
#> FALSE TRUE
#> 0.251 0.749
#>
#> [[1]]$O1
#> A B C D E
#> 0 0 0 0 1
#>
#> [[1]]$O2
#> A B C D E
#> 0 1 0 0 0
#>
#> [[1]]$O3
#> A B C D E
#> 1 0 0 0 0
#>
#> [[1]]$O4
#> A B C D E
#> 1 0 0 0 0
#>
#> [[1]]$O5
#> A B C D E
#> 0 0 1 0 0
# compare with the observed and true data
rbind(truth = dat$truth_obs[2L, ], observed = dat$seen_obs[2L, ])
#> C1 C2 C3 C4 C5 B1 B2 B3 B4 B5 O1 O2 O3
#> truth 0.142 2.63 0.338 0.0927 0.000152 FALSE TRUE TRUE FALSE TRUE E B A
#> observed 0.142 NA NA 0.0927 0.000152 FALSE NA TRUE NA NA E B A
#> O4 O5
#> truth A C
#> observed A C
# we can threshold the data like this
threshold <- function(org_data, imputed){
# checks
stopifnot(NROW(org_data) == length(imputed),
is.list(imputed), is.data.frame(org_data))
# threshold
is_cont <- which(sapply(org_data, is.numeric))
is_bin <- which(sapply(org_data, is.logical))
is_ord <- which(sapply(org_data, is.ordered))
stopifnot(
length(is_cont) + length(is_bin) + length(is_ord) == NCOL(org_data))
is_cat <- c(is_bin, is_ord)
trans_to_df <- function(x){
if(is.matrix(x))
as.data.frame(t(x))
else
as.data.frame( x )
}
out_cont <- trans_to_df(sapply(imputed, function(x) unlist(x[is_cont])))
out_cat <- trans_to_df(sapply(imputed, function(x)
sapply(x[is_cat], which.max)))
out <- cbind(out_cont, out_cat)
# set factor levels etc.
out <- out[, order(c(is_cont, is_bin, is_ord))]
if(length(is_bin) > 0)
out[, is_bin] <- out[, is_bin] > 1L
if(length(is_ord) > 0)
for(i in is_ord)
out[[i]] <- ordered(
unlist(out[[i]]), labels = levels(org_data[, i]))
colnames(out) <- colnames(org_data)
out
}
thresh_dat <- threshold(dat$seen_obs, imp_res)
# compare thresholded data with observed and true data
head(thresh_dat)
#> C1 C2 C3 C4 C5 B1 B2 B3 B4 B5 O1 O2 O3 O4
#> 1 0.237 0.693 0.798 0.0666 1.216658 FALSE FALSE FALSE FALSE TRUE E C B B
#> 2 0.142 2.083 0.249 0.0927 0.000152 FALSE TRUE TRUE FALSE TRUE E B A A
#> 3 1.301 0.748 0.629 0.4280 0.572448 FALSE TRUE TRUE TRUE TRUE E A E C
#> 4 2.702 0.775 1.128 2.1776 1.700870 FALSE TRUE TRUE TRUE TRUE A B D B
#> 5 0.925 0.908 0.205 0.6046 0.171311 TRUE TRUE FALSE FALSE FALSE E B D D
#> 6 0.115 1.063 1.341 0.2399 0.238368 FALSE TRUE TRUE FALSE TRUE E A A B
#> O5
#> 1 D
#> 2 C
#> 3 E
#> 4 E
#> 5 E
#> 6 B
head(dat$seen_obs) # observed data
#> C1 C2 C3 C4 C5 B1 B2 B3 B4 B5 O1 O2
#> 1 0.237 0.693 0.798 0.0666 NA FALSE FALSE FALSE FALSE TRUE E C
#> 2 0.142 NA NA 0.0927 0.000152 FALSE NA TRUE NA NA E B
#> 3 NA 0.748 0.629 0.4280 NA NA TRUE NA NA TRUE <NA> A
#> 4 2.702 NA NA 2.1776 1.700870 FALSE TRUE TRUE NA TRUE A <NA>
#> 5 0.925 NA 0.205 0.6046 0.171311 TRUE TRUE FALSE FALSE FALSE E B
#> 6 0.115 NA 1.341 NA NA FALSE TRUE TRUE NA NA E <NA>
#> O3 O4 O5
#> 1 B B <NA>
#> 2 A A C
#> 3 <NA> C E
#> 4 D B <NA>
#> 5 <NA> D <NA>
#> 6 A B <NA>
head(dat$truth_obs) # true data
#> C1 C2 C3 C4 C5 B1 B2 B3 B4 B5 O1 O2 O3 O4
#> 1 0.237 0.693 0.798 0.0666 0.950476 FALSE FALSE FALSE FALSE TRUE E C B B
#> 2 0.142 2.630 0.338 0.0927 0.000152 FALSE TRUE TRUE FALSE TRUE E B A A
#> 3 2.864 0.748 0.629 0.4280 1.341650 FALSE TRUE TRUE FALSE TRUE C A D C
#> 4 2.702 1.153 0.459 2.1776 1.700870 FALSE TRUE TRUE TRUE TRUE A C D B
#> 5 0.925 0.365 0.205 0.6046 0.171311 TRUE TRUE FALSE FALSE FALSE E B B D
#> 6 0.115 0.563 1.341 0.7184 0.306274 FALSE TRUE TRUE FALSE TRUE E A A B
#> O5
#> 1 E
#> 2 C
#> 3 E
#> 4 E
#> 5 E
#> 6 B
# compare correct categories
get_classif_error <- function(impu_dat, truth = dat$truth_obs,
observed = dat$seen_obs){
is_cat <- sapply(truth, function(x)
is.logical(x) || is.ordered(x))
is_match <- impu_dat[, is_cat] == truth[, is_cat]
is_match[!is.na(observed[, is_cat])] <- NA_integer_
1 - colMeans(is_match, na.rm = TRUE)
}
get_classif_error(thresh_dat)
#> B1 B2 B3 B4 B5 O1 O2 O3 O4 O5
#> 0.274 0.295 0.226 0.320 0.288 0.566 0.638 0.616 0.600 0.548
# compute RMSE
get_rmse <- function(impu_dat, truth = dat$truth_obs,
observed = dat$seen_obs){
is_con <- sapply(truth, is.numeric)
err <- as.matrix(impu_dat[, is_con] - truth[, is_con])
err[!is.na(observed[, is_con])] <- NA_real_
sqrt(colMeans(err^2, na.rm = TRUE))
}
get_rmse(thresh_dat)
#> C1 C2 C3 C4 C5
#> 0.644 0.784 0.652 0.795 0.746
# we can compare this with missForest
miss_forest_arg <- dat$seen_obs
is_log <- sapply(miss_forest_arg, is.logical)
miss_forest_arg[, is_log] <- lapply(miss_forest_arg[, is_log], as.factor)
set.seed(1)
system.time(miss_res <- missForest(miss_forest_arg))
#> missForest iteration 1 in progress...done!
#> missForest iteration 2 in progress...done!
#> missForest iteration 3 in progress...done!
#> missForest iteration 4 in progress...done!
#> missForest iteration 5 in progress...done!
#> missForest iteration 6 in progress...done!
#> missForest iteration 7 in progress...done!
#> missForest iteration 8 in progress...done!
#> missForest iteration 9 in progress...done!
#> user system elapsed
#> 44.58 0.06 44.64
# turn binary variables back to logicals
miss_res$ximp[, is_log] <- lapply(
miss_res$ximp[, is_log], function(x) as.integer(x) > 1L)
rbind(mdgc = get_classif_error(thresh_dat),
missForest = get_classif_error(miss_res$ximp))
#> B1 B2 B3 B4 B5 O1 O2 O3 O4 O5
#> mdgc 0.274 0.295 0.226 0.320 0.288 0.566 0.638 0.616 0.600 0.548
#> missForest 0.315 0.340 0.304 0.371 0.319 0.651 0.726 0.680 0.673 0.612
rbind(mdgc = get_rmse(thresh_dat),
missForest = get_rmse(miss_res$ximp))
#> C1 C2 C3 C4 C5
#> mdgc 0.644 0.784 0.652 0.795 0.746
#> missForest 0.806 0.848 0.755 0.845 0.842Here is an example where we use the mdgc function to do the model
estimation and the imputation:
# have a data set with missing continuous, binary, and ordinal variables
head(dat$seen_obs)
#> C1 C2 C3 C4 C5 B1 B2 B3 B4 B5 O1 O2
#> 1 0.237 0.693 0.798 0.0666 NA FALSE FALSE FALSE FALSE TRUE E C
#> 2 0.142 NA NA 0.0927 0.000152 FALSE NA TRUE NA NA E B
#> 3 NA 0.748 0.629 0.4280 NA NA TRUE NA NA TRUE <NA> A
#> 4 2.702 NA NA 2.1776 1.700870 FALSE TRUE TRUE NA TRUE A <NA>
#> 5 0.925 NA 0.205 0.6046 0.171311 TRUE TRUE FALSE FALSE FALSE E B
#> 6 0.115 NA 1.341 NA NA FALSE TRUE TRUE NA NA E <NA>
#> O3 O4 O5
#> 1 B B <NA>
#> 2 A A C
#> 3 <NA> C E
#> 4 D B <NA>
#> 5 <NA> D <NA>
#> 6 A B <NA>
# perform the estimation and imputation
set.seed(1)
system.time(res <- mdgc(dat$seen_obs, verbose = TRUE, maxpts = 5000L,
n_threads = 4L, maxit = 25L, use_aprx = TRUE))
#> Estimating the model...
#> End of iteration 1 with learning rate 0.00100000
#> Log marginal likelihood approximation is -23441.64
#> Previous approximate gradient norm was 3444.23
#>
#> End of iteration 2 with learning rate 0.00098000
#> Log marginal likelihood approximation is -23389.38
#> Previous approximate gradient norm was 1782.50
#>
#> End of iteration 3 with learning rate 0.00096040
#> Log marginal likelihood approximation is -23366.99
#> Previous approximate gradient norm was 1157.05
#>
#> End of iteration 4 with learning rate 0.00094119
#> Log marginal likelihood approximation is -23355.16
#> Previous approximate gradient norm was 849.85
#>
#> End of iteration 5 with learning rate 0.00092237
#> Log marginal likelihood approximation is -23348.19
#> Previous approximate gradient norm was 667.85
#>
#> End of iteration 6 with learning rate 0.00090392
#> Log marginal likelihood approximation is -23343.80
#> Previous approximate gradient norm was 548.25
#>
#> End of iteration 7 with learning rate 0.00088584
#> Log marginal likelihood approximation is -23340.81
#> Previous approximate gradient norm was 469.87
#>
#> End of iteration 8 with learning rate 0.00086813
#> Log marginal likelihood approximation is -23338.67
#> Previous approximate gradient norm was 412.60
#>
#> End of iteration 9 with learning rate 0.00085076
#> Log marginal likelihood approximation is -23337.12
#> Previous approximate gradient norm was 370.90
#>
#> End of iteration 10 with learning rate 0.00083375
#> Log marginal likelihood approximation is -23335.95
#> Previous approximate gradient norm was 337.75
#>
#> End of iteration 11 with learning rate 0.00081707
#> Log marginal likelihood approximation is -23335.04
#> Previous approximate gradient norm was 315.68
#>
#> End of iteration 12 with learning rate 0.00080073
#> Log marginal likelihood approximation is -23334.32
#> Previous approximate gradient norm was 296.75
#>
#> End of iteration 13 with learning rate 0.00078472
#> Log marginal likelihood approximation is -23333.75
#> Previous approximate gradient norm was 281.62
#>
#> End of iteration 14 with learning rate 0.00076902
#> Log marginal likelihood approximation is -23333.27
#> Previous approximate gradient norm was 268.38
#>
#> Performing imputation...
#> user system elapsed
#> 11.91 0.00 3.43
# compare the estimated correlation matrix with the truth
norm(dat$Sigma - res$vcov, "F") / norm(dat$Sigma, "F")
#> [1] 0.0956
# compute the classifcation error and RMSE
get_classif_error(res$ximp)
#> B1 B2 B3 B4 B5 O1 O2 O3 O4 O5
#> 0.272 0.297 0.225 0.318 0.289 0.576 0.637 0.626 0.602 0.541
get_rmse(res$ximp)
#> C1 C2 C3 C4 C5
#> 0.644 0.784 0.652 0.795 0.746We can compare this with the mixedgcImp which uses the method
described in Zhao and Udell (2020b):
# turn the data to a format that can be based
dat_pass <- dat$seen_obs
is_cat <- sapply(dat_pass, function(x) is.logical(x) | is.ordered(x))
dat_pass[, is_cat] <- lapply(dat_pass[, is_cat], as.integer)
system.time(imp_apr_em <- impute_mixedgc(dat_pass, eps = 1e-4))
#> user system elapsed
#> 20 0 20
# compare the estimated correlation matrix with the truth
get_rel_err <- function(est, keep = seq_len(NROW(truth)), truth = dat$Sigma)
norm(truth[keep, keep] - est[keep, keep], "F") /
norm(truth, "F")
c(mdgc = get_rel_err(res$vcov),
mixedgcImp = get_rel_err(imp_apr_em$R),
`mdgc bin/ordered` = get_rel_err(res$vcov , is_cat),
`mixedgcImp bin/ordered` = get_rel_err(imp_apr_em$R, is_cat),
`mdgc continuous` = get_rel_err(res$vcov , !is_cat),
`mixedgcImp continuous` = get_rel_err(imp_apr_em$R, !is_cat))
#> mdgc mixedgcImp mdgc bin/ordered
#> 0.0956 0.1243 0.0743
#> mixedgcImp bin/ordered mdgc continuous mixedgcImp continuous
#> 0.1083 0.0242 0.0257
# compare the classifcation error and RMSE
imp_apr_res <- as.data.frame(imp_apr_em$Ximp)
is_bin <- sapply(dat$seen_obs, is.logical)
imp_apr_res[, is_bin] <- lapply(imp_apr_res[, is_bin], `>`, e2 = 0)
is_ord <- sapply(dat$seen_obs, is.ordered)
imp_apr_res[, is_ord] <- mapply(function(x, idx)
ordered(x, labels = levels(dat$seen_obs[[idx]])),
x = imp_apr_res[, is_ord], i = which(is_ord), SIMPLIFY = FALSE)
rbind(mdgc = get_classif_error(res$ximp),
mixedgcImp = get_classif_error(imp_apr_res))
#> B1 B2 B3 B4 B5 O1 O2 O3 O4 O5
#> mdgc 0.272 0.297 0.225 0.318 0.289 0.576 0.637 0.626 0.602 0.541
#> mixedgcImp 0.281 0.328 0.232 0.320 0.288 0.626 0.694 0.688 0.609 0.556
rbind(mdgc = get_rmse(res$ximp),
mixedgcImp = get_rmse(imp_apr_res))
#> C1 C2 C3 C4 C5
#> mdgc 0.644 0.784 0.652 0.795 0.746
#> mixedgcImp 0.645 0.789 0.655 0.810 0.755We will perform a simulation study in this section to compare different methods in terms of their computation time and performance. We first perform the simulation.
# the seeds we will use
seeds <- c(293498804L, 311878062L, 370718465L, 577520465L, 336825034L, 661670751L, 750947065L, 255824398L, 281823005L, 721186455L, 251974931L, 643568686L, 273097364L, 328663824L, 490259480L, 517126385L, 651705963L, 43381670L, 503505882L, 609251792L, 643001919L, 244401725L, 983414550L, 850590318L, 714971434L, 469416928L, 237089923L, 131313381L, 689679752L, 344399119L, 330840537L, 6287534L, 735760574L, 477353355L, 579527946L, 83409653L, 710142087L, 830103443L, 94094987L, 422058348L, 117889526L, 259750108L, 180244429L, 762680168L, 112163383L, 10802048L, 440434442L, 747282444L, 736836365L, 837303896L, 50697895L, 231661028L, 872653438L, 297024405L, 719108161L, 201103881L, 485890767L, 852715172L, 542126886L, 155221223L, 18987375L, 203133067L, 460377933L, 949381283L, 589083178L, 820719063L, 543339683L, 154667703L, 480316186L, 310795921L, 287317945L, 30587393L, 381290126L, 178269809L, 939854883L, 660119506L, 825302990L, 764135140L, 433746745L, 173637986L, 100446967L, 333304121L, 225525537L, 443031789L, 587486506L, 245392609L, 469144801L, 44073812L, 462948652L, 226692940L, 165285895L, 546908869L, 550076645L, 872290900L, 452044364L, 620131127L, 600097817L, 787537854L, 15915195L, 64220696L)
# gather or compute the results (you may skip this)
res <- lapply(seeds, function(s){
file_name <- file.path("sim-res", sprintf("seed-%d.RDS", s))
if(file.exists(file_name)){
message(sprintf("Reading '%s'", file_name))
out <- readRDS(file_name)
} else {
message(sprintf("Running '%s'", file_name))
# simulate the data
set.seed(s)
dat <- sim_dat(2000L, p = 15L)
# fit models and impute
mdgc_time <- system.time(
mdgc_res <- mdgc(dat$seen_obs, verbose = FALSE, maxpts = 5000L,
n_threads = 4L, maxit = 25L, use_aprx = TRUE))
dat_pass <- dat$seen_obs
is_cat <- sapply(dat_pass, function(x) is.logical(x) | is.ordered(x))
dat_pass[, is_cat] <- lapply(dat_pass[, is_cat], as.integer)
mixedgc_time <-
system.time(mixedgc_res <- impute_mixedgc(dat_pass, eps = 1e-4))
miss_forest_arg <- dat$seen_obs
is_log <- sapply(miss_forest_arg, is.logical)
miss_forest_arg[, is_log] <- lapply(
miss_forest_arg[, is_log], as.factor)
sink(tempfile())
miss_time <- system.time(
miss_res <- missForest(miss_forest_arg, verbose = FALSE))
sink()
miss_res$ximp[, is_log] <- lapply(
miss_res$ximp[, is_log], function(x) as.integer(x) > 1L)
# impute using the other estimate
mdgc_obj <- get_mdgc(dat$seen_obs)
impu_mixedgc_est <- mdgc_impute(mdgc_obj, mixedgc_res$R, mdgc_obj$means)
impu_mixedgc_est <- threshold(dat$seen_obs, impu_mixedgc_est)
# gather output for the correlation matrix estimates
vcov_res <- list(truth = dat$Sigma, mdgc = mdgc_res$vcov,
mixedgc = mixedgc_res$R)
get_rel_err <- function(est, truth, keep = seq_len(NROW(truth)))
norm(truth[keep, keep] - est[keep, keep], "F") / norm(truth, "F")
vcov_res <- within(vcov_res, {
mdgc_rel_err = get_rel_err(mdgc , truth)
mixedgc_rel_err = get_rel_err(mixedgc, truth)
})
# gather the estimated means
mea_ests <- list(marginal = mdgc_obj$means,
joint = mdgc_res$mea)
# gather output for the imputation
mixedgc_imp_res <- as.data.frame(mixedgc_res$Ximp)
is_bin <- sapply(dat$seen_obs, is.logical)
mixedgc_imp_res[, is_bin] <-
lapply(mixedgc_imp_res[, is_bin, drop = FALSE], `>`, e2 = 0)
is_ord <- sapply(dat$seen_obs, is.ordered)
mixedgc_imp_res[, is_ord] <- mapply(function(x, idx)
ordered(x, labels = levels(dat$seen_obs[[idx]])),
x = mixedgc_imp_res[, is_ord, drop = FALSE],
i = which(is_ord), SIMPLIFY = FALSE)
get_bin_err <- function(x){
. <- function(z) z[, is_bin, drop = FALSE]
get_classif_error(
.(x), truth = .(dat$truth_obs), observed = .(dat$seen_obs))
}
get_ord_err <- function(x){
. <- function(z) z[, is_ord, drop = FALSE]
get_classif_error(
.(x), truth = .(dat$truth_obs), observed = .(dat$seen_obs))
}
err <- list(
mdgc_bin = get_bin_err(mdgc_res$ximp),
mixedgc_bin = get_bin_err(mixedgc_imp_res),
mixed_bin = get_bin_err(impu_mixedgc_est),
missForest_bin = get_bin_err(miss_res$ximp),
mdgc_class = get_ord_err(mdgc_res$ximp),
mixedgc_class = get_ord_err(mixedgc_imp_res),
mixed_class = get_ord_err(impu_mixedgc_est),
missForest_class = get_ord_err(miss_res$ximp),
mdgc_rmse = get_rmse(
mdgc_res$ximp, truth = dat$truth_obs, observed = dat$seen_obs),
mixedgc_rmse = get_rmse(
mixedgc_imp_res, truth = dat$truth_obs, observed = dat$seen_obs),
mixed_rmse = get_rmse(
impu_mixedgc_est, truth = dat$truth_obs, observed = dat$seen_obs),
missForest_rmse = get_rmse(
miss_res$ximp, truth = dat$truth_obs, observed = dat$seen_obs))
# gather the times
times <- list(mdgc = mdgc_time, mixedgc = mixedgc_time,
missForest = miss_time)
# save stats to check convergence
conv_stats <- list(mdgc = mdgc_res$logLik,
mixedgc = mixedgc_res$loglik)
# save output
out <- list(vcov_res = vcov_res, err = err, times = times,
conv_stats = conv_stats, mea_ests = mea_ests)
saveRDS(out, file_name)
}
# print summary stat to the console while knitting
out <- readRDS(file_name)
. <- function(x)
message(paste(sprintf("%8.3f", x), collapse = " "))
with(out, {
message(paste(
"mdgc logLik",
paste(sprintf("%.2f", conv_stats$mdgc), collapse = " ")))
message(paste(
"mixedgc logLik",
paste(sprintf("%.2f", conv_stats$mixedgc), collapse = " ")))
message(sprintf(
"Relative correlation matrix estimate errors are %.4f %.4f",
vcov_res$mdgc_rel_err, vcov_res$mixedgc_rel_err))
message(sprintf(
"Times are %.2f %.2f %.2f",
times$mdgc["elapsed"], times$mixedgc["elapsed"],
times$missForest["elapsed"]))
message(sprintf(
"Binary classifcation errors are %.2f %.2f %.2f (%.2f)",
mean(err$mdgc_bin), mean(err$mixedgc_bin),
mean(err$missForest_bin), mean(err$mixed_bin)))
message(sprintf(
"Ordinal classifcation errors are %.2f %.2f %.2f (%.2f)",
mean(err$mdgc_class), mean(err$mixedgc_class),
mean(err$missForest_class), mean(err$mixed_class)))
message(sprintf(
"Mean RMSEs are %.2f %.2f %.2f (%.2f)",
mean(err$mdgc_rmse), mean(err$mixedgc_rmse),
mean(err$missForest_rmse), mean(err$mixed_rmse)))
message("")
})
out
})The difference in computation time is given below:
# assign function to show the summary stats
show_sim_stats <- function(v1, v2, v3, what, sub_ele = NULL){
vals <- sapply(res, function(x)
do.call(rbind, x[[what]][c(v1, v2, v3)]),
simplify = "array")
if(!is.null(sub_ele))
vals <- vals[, sub_ele, , drop = FALSE]
cat("Means and standard errors:\n")
mea_se <- function(x)
c(mean = mean(x), SE = sd(x) / sqrt(length(x)))
print(t(apply(vals, 1L, mea_se)))
cat("\nDifference:\n")
print(t(apply(
c(vals[v1, , ]) -
aperm(vals[c(v2, v3), , , drop = FALSE], c(3L, 2L, 1L)),
3L, mea_se)))
}
# compare estimation time
show_sim_stats(1L, 2L, 3L, "times", "elapsed")
#> Means and standard errors:
#> mean SE
#> mdgc 3.68 0.0648
#> mixedgc 21.06 0.1455
#> missForest 46.71 0.6242
#>
#> Difference:
#> mean SE
#> mixedgc -17.4 0.155
#> missForest -43.0 0.620The summary stats for the relative Frobenius norm between the estimated and true correlation matrix is given below:
# relative norms
show_sim_stats("mixedgc_rel_err", "mdgc_rel_err", NULL, "vcov_res")
#> Means and standard errors:
#> mean SE
#> mixedgc_rel_err 0.1187 0.001230
#> mdgc_rel_err 0.0867 0.000972
#>
#> Difference:
#> mean SE
#> mdgc_rel_err 0.032 0.000909Finally, here are the results for the classification error for the binary and ordinal outcomes and the root mean square error:
# the binary variables
show_sim_stats("mdgc_bin", "mixedgc_bin", "missForest_bin", "err")
#> Means and standard errors:
#> mean SE
#> mdgc_bin 0.244 0.00187
#> mixedgc_bin 0.252 0.00186
#> missForest_bin 0.294 0.00188
#>
#> Difference:
#> mean SE
#> mixedgc_bin -0.0074 0.00255
#> missForest_bin -0.0500 0.00255
# the ordinal variables
show_sim_stats("mdgc_class", "mixedgc_class", "missForest_class", "err")
#> Means and standard errors:
#> mean SE
#> mdgc_class 0.590 0.00215
#> mixedgc_class 0.623 0.00245
#> missForest_class 0.658 0.00173
#>
#> Difference:
#> mean SE
#> mixedgc_class -0.0332 0.00317
#> missForest_class -0.0680 0.00272
# the continuous variables
show_sim_stats("mdgc_rmse", "mixedgc_rmse", "missForest_rmse", "err")
#> Means and standard errors:
#> mean SE
#> mdgc_rmse 0.760 0.00402
#> mixedgc_rmse 0.767 0.00404
#> missForest_rmse 0.850 0.00340
#>
#> Difference:
#> mean SE
#> mixedgc_rmse -0.00738 0.00575
#> missForest_rmse -0.09016 0.00525It is important to emphasize that missForest is not estimating the true model.
We extend the model suggested by Zhao and Udell (2020b) in this section. The example is very similar to the previous one but with multinomial variables.
# simulates a data set and mask some of the data.
#
# Args:
# n: number of observations.
# p: number of variables.
# n_lvls: number of levels for the ordinal and multinomial variables.
# verbose: print status during the simulation.
#
# Returns:
# Simulated masked data, the true data, and true covariance matrix.
sim_dat <- function(n, p = 4L, n_lvls = 5L, verbose = FALSE){
# determine the type
n_rep <- floor((p + 4 - 1) / 4)
type <- rep(1:4, n_rep)[1:p]
is_con <- type == 1L
is_bin <- type == 2L
is_ord <- type == 3L
is_mult <- type == 4L
col_nam <- c(outer(c("C", "B", "O", "M"), 1:n_rep, paste0))[1:p]
idx <- head(cumsum(c(1L, ifelse(type == 4, n_lvls, 1L))), -1L)
# get the covariance matrix
n_latent <- p + (n_lvls - 1L) * (p %/% 4)
Sig <- drop(rWishart(1L, 2 * n_latent, diag(1 / n_latent / 2, n_latent)))
# essentially set the reference level to zero
for(i in idx[is_mult]){
Sig[i, ] <- 0
Sig[ , i] <- 0
}
# rescale some rows and columns
sds <- sqrt(diag(Sig))
for(i in idx[is_mult]){
sds[i] <- 1
sds[i + 3:n_lvls - 1] <- 1
}
Sig <- diag(1/sds) %*% Sig %*% diag(1/sds)
# draw the observations
truth <- mvtnorm::rmvnorm(n, sigma = Sig)
truth[, idx[is_mult]] <- 0
# sample which are masked data
is_mask <- matrix(runif(n * p) < .3, n)
# make sure we have no rows with all missing data
while(any(all_nans <- rowSums(is_mask) == NCOL(is_mask)))
is_mask[all_nans, ] <- runif(sum(all_nans) * p) < .3
# create the observed data
truth_obs <- lapply(type, function(i) if(i == 1L) numeric(n) else integer(n))
truth_obs <- data.frame(truth_obs)
colnames(truth_obs) <- col_nam
bs_ord <- qnorm(seq(0, 1, length.out = n_lvls + 1L))
for(i in 1:p){
idx_i <- idx[i]
switch(
type[i],
# continuous
truth_obs[, i] <- qexp(pnorm(truth[, idx_i])),
# binary
truth_obs[, i] <- truth[, idx_i] > 0,
# ordinal
{
truth_obs[, i] <-
ordered(as.integer(cut(truth[, idx_i], breaks = bs_ord)))
levels(truth_obs[, i]) <-
LETTERS[seq_len(length(unique(truth_obs[, i])))]
},
# multinomial
{
truth_obs[, i] <- apply(
truth[, idx_i + 1:n_lvls - 1L], 1L, which.max)
truth_obs[, i] <- factor(truth_obs[, i],
labels = paste0("T", 1:n_lvls))
},
stop("Type is not implemented"))
}
# mask the data
seen_obs <- truth_obs
seen_obs[is_mask] <- NA
list(truth = truth, truth_obs = truth_obs, seen_obs = seen_obs,
Sigma = Sig)
}
# simulate and show the data
set.seed(1)
p <- 8L
dat <- sim_dat(2000L, p = p, verbose = TRUE, n_lvls = 4)
# show the first rows of the observed data
head(dat$seen_obs)
#> C1 B1 O1 M1 C2 B2 O2 M2
#> 1 NA TRUE B T1 1.989 FALSE C T2
#> 2 0.206 NA A T2 NA TRUE B T3
#> 3 0.111 FALSE B T2 0.132 NA <NA> T3
#> 4 NA NA <NA> T2 1.137 TRUE <NA> T3
#> 5 NA TRUE <NA> T4 NA FALSE B <NA>
#> 6 0.320 FALSE <NA> T3 0.492 FALSE A T2
# assign object to perform the estimation and the imputation
obj <- get_mdgc(dat$seen_obs)
ptr <- get_mdgc_log_ml(obj)
# get starting values
start_vals <- mdgc_start_value(obj)
# plot the starting values and the true values
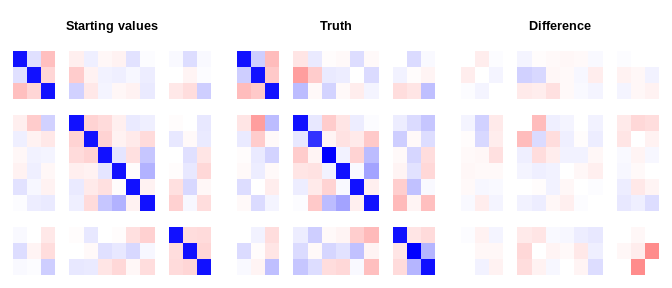
do_plot <- function(est, truth, main){
par_old <- par(mfcol = c(1, 3), mar = c(1, 1, 4, 1))
on.exit(par(par_old))
sc <- colorRampPalette(c("Red", "White", "Blue"))(201)
ma <- max(abs(est), max(abs(truth)))
f <- function(x, main)
image(x[, NCOL(x):1], main = main, col = sc, zlim = c(-ma, ma),
xaxt = "n", yaxt = "n", bty = "n")
f(est, main)
f(truth, "Truth")
f(est - truth, "Difference")
}
do_plot(start_vals, dat$Sigma, "Starting values")# check the log marginal likelihood at the starting values and compare with
# the true values at the starting values
mdgc_log_ml(ptr, start_vals, mea = obj$means, n_threads = 1L)
#> [1] -13185
# and at the true values
mdgc_log_ml(ptr, dat$Sigma , mea = numeric(length(obj$means)),
n_threads = 1L)
#> [1] -13140
# much better than using a diagonal matrix!
mdgc_log_ml(ptr, diag(NROW(dat$Sigma)), mea = obj$means, n_threads = 1L)
#> [1] -13622
# estimate the model
system.time(
ests <- mdgc_fit(ptr, vcov = start_vals, mea = obj$means,
method = "aug_Lagran",
n_threads = 4L, rel_eps = 1e-2, maxpts = 1000L,
minvls = 200L, use_aprx = TRUE, conv_crit = 1e-8))
#> user system elapsed
#> 196.8 0.0 49.2
# refine the estimates
system.time(
ests <- mdgc_fit(ptr, vcov = ests$result$vcov,
mea = ests$result$mea,
method = "aug_Lagran",
n_threads = 4L, rel_eps = 1e-3, maxpts = 10000L,
minvls = 1000L, mu = ests$mu, lambda = ests$lambda,
use_aprx = TRUE, conv_crit = 1e-8))
#> user system elapsed
#> 498 0 128
# use ADAM
system.time(
fit_adam <- mdgc_fit(
ptr, vcov = start_vals, mea = obj$means, minvls = 200L,
n_threads = 4L, lr = 1e-3, maxit = 25L, batch_size = 100L,
method = "adam", rel_eps = 1e-3, maxpts = 5000L,
use_aprx = TRUE))
#> user system elapsed
#> 18.98 0.00 4.75
# use SVRG
system.time(
fit_svrg <- mdgc_fit(
ptr, vcov = start_vals, mea = obj$means, minvls = 200L,
n_threads = 4L, lr = 1e-3, maxit = 25L, batch_size = 100L,
method = "svrg", verbose = TRUE, rel_eps = 1e-3, maxpts = 5000L,
use_aprx = TRUE, conv_crit = 1e-8))
#> End of iteration 1 with learning rate 0.00100000
#> Log marginal likelihood approximation is -13162.37
#> Previous approximate gradient norm was 1982.96
#>
#> End of iteration 2 with learning rate 0.00098000
#> Log marginal likelihood approximation is -13135.74
#> Previous approximate gradient norm was 1176.34
#>
#> End of iteration 3 with learning rate 0.00096040
#> Log marginal likelihood approximation is -13125.17
#> Previous approximate gradient norm was 762.23
#>
#> End of iteration 4 with learning rate 0.00094119
#> Log marginal likelihood approximation is -13119.99
#> Previous approximate gradient norm was 529.22
#>
#> End of iteration 5 with learning rate 0.00092237
#> Log marginal likelihood approximation is -13117.07
#> Previous approximate gradient norm was 390.78
#>
#> End of iteration 6 with learning rate 0.00090392
#> Log marginal likelihood approximation is -13115.27
#> Previous approximate gradient norm was 307.94
#>
#> End of iteration 7 with learning rate 0.00088584
#> Log marginal likelihood approximation is -13114.08
#> Previous approximate gradient norm was 254.53
#>
#> End of iteration 8 with learning rate 0.00086813
#> Log marginal likelihood approximation is -13113.28
#> Previous approximate gradient norm was 216.15
#>
#> End of iteration 9 with learning rate 0.00085076
#> Log marginal likelihood approximation is -13112.71
#> Previous approximate gradient norm was 186.11
#>
#> End of iteration 10 with learning rate 0.00083375
#> Log marginal likelihood approximation is -13112.29
#> Previous approximate gradient norm was 163.70
#>
#> End of iteration 11 with learning rate 0.00081707
#> Log marginal likelihood approximation is -13111.99
#> Previous approximate gradient norm was 147.40
#>
#> End of iteration 12 with learning rate 0.00080073
#> Log marginal likelihood approximation is -13111.77
#> Previous approximate gradient norm was 133.51
#>
#> End of iteration 13 with learning rate 0.00078472
#> Log marginal likelihood approximation is -13111.59
#> Previous approximate gradient norm was 121.95
#>
#> End of iteration 14 with learning rate 0.00076902
#> Log marginal likelihood approximation is -13111.45
#> Previous approximate gradient norm was 111.37
#>
#> End of iteration 15 with learning rate 0.00075364
#> Log marginal likelihood approximation is -13111.37
#> Previous approximate gradient norm was 116.29
#>
#> End of iteration 16 with learning rate 0.00073857
#> Log marginal likelihood approximation is -13111.26
#> Previous approximate gradient norm was 108.67
#>
#> End of iteration 17 with learning rate 0.00072380
#> Log marginal likelihood approximation is -13111.18
#> Previous approximate gradient norm was 102.82
#>
#> End of iteration 18 with learning rate 0.00070932
#> Log marginal likelihood approximation is -13111.11
#> Previous approximate gradient norm was 98.91
#>
#> End of iteration 19 with learning rate 0.00069514
#> Log marginal likelihood approximation is -13111.04
#> Previous approximate gradient norm was 95.78
#>
#> End of iteration 20 with learning rate 0.00068123
#> Log marginal likelihood approximation is -13111.00
#> Previous approximate gradient norm was 91.77
#>
#> End of iteration 21 with learning rate 0.00066761
#> Log marginal likelihood approximation is -13110.96
#> Previous approximate gradient norm was 88.01
#>
#> End of iteration 22 with learning rate 0.00065426
#> Log marginal likelihood approximation is -13110.94
#> Previous approximate gradient norm was 85.42
#>
#> End of iteration 23 with learning rate 0.00064117
#> Log marginal likelihood approximation is -13110.90
#> Previous approximate gradient norm was 83.17
#>
#> End of iteration 24 with learning rate 0.00062835
#> Log marginal likelihood approximation is -13110.84
#> Previous approximate gradient norm was 83.19
#>
#> End of iteration 25 with learning rate 0.00061578
#> Log marginal likelihood approximation is -13110.82
#> Previous approximate gradient norm was 80.33
#> user system elapsed
#> 38.379 0.004 9.597
# compare log marginal likelihood
print(rbind(
`Augmented Lagrangian` =
mdgc_log_ml(ptr, ests$result$vcov , mea = ests$result$mea,
n_threads = 1L),
ADAM =
mdgc_log_ml(ptr, fit_adam$result$vcov, mea = fit_adam$result$mea,
n_threads = 1L),
SVRG =
mdgc_log_ml(ptr, fit_svrg$result$vcov, mea = fit_svrg$result$mea,
n_threads = 1L),
Truth =
mdgc_log_ml(ptr, dat$Sigma , mea = numeric(length(obj$means)),
n_threads = 1L)), digits = 10)
#> [,1]
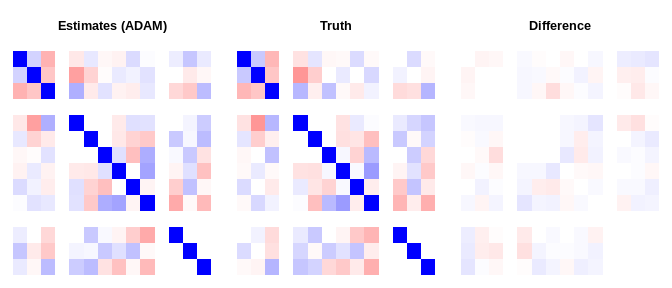
#> Augmented Lagrangian -13111.01596
#> ADAM -13112.96534
#> SVRG -13110.74984
#> Truth -13140.31880
# compare the estimated and the true values (should not match because of
# overparameterization? See https://stats.stackexchange.com/q/504682/81865)
do_plot(ests$result$vcov , dat$Sigma, "Estimates (Aug. Lagrangian)")do_plot(fit_adam$result$vcov, dat$Sigma, "Estimates (ADAM)")do_plot(fit_svrg$result$vcov, dat$Sigma, "Estimates (SVRG)")# after rescaling
do_plot_rescale <- function(x, lab){
trans <- function(z){
scal <- diag(NCOL(z))
m <- obj$multinomial[[1L]]
for(i in seq_len(NCOL(m))){
idx <- m[3, i] + 1 + seq_len(m[2, i] - 1)
scal[idx, idx] <- solve(t(chol(z[idx, idx])))
}
tcrossprod(scal %*% z, scal)
}
do_plot(trans(x), trans(dat$Sigma), lab)
}
do_plot_rescale(ests$result$vcov , "Estimates (Aug. Lagrangian)")do_plot_rescale(fit_adam$result$vcov, "Estimates (ADAM)")do_plot_rescale(fit_svrg$result$vcov, "Estimates (SVRG)")# perform the imputation
system.time(
imp_res <- mdgc_impute(obj, ests$result$vcov, mea = ests$result$mea,
rel_eps = 1e-3, maxit = 10000L, n_threads = 4L))
#> user system elapsed
#> 11.99 0.00 3.19
# look at the result for one of the observations
imp_res[1L]
#> [[1]]
#> [[1]]$C1
#> [1] 0.714
#>
#> [[1]]$B1
#> FALSE TRUE
#> 0 1
#>
#> [[1]]$O1
#> A B C D
#> 0 1 0 0
#>
#> [[1]]$M1
#> T1 T2 T3 T4
#> 1 0 0 0
#>
#> [[1]]$C2
#> [1] 1.99
#>
#> [[1]]$B2
#> FALSE TRUE
#> 1 0
#>
#> [[1]]$O2
#> A B C D
#> 0 0 1 0
#>
#> [[1]]$M2
#> T1 T2 T3 T4
#> 0 1 0 0
# compare with the observed and true data
rbind(truth = dat$truth_obs[1L, ], observed = dat$seen_obs[1L, ])
#> C1 B1 O1 M1 C2 B2 O2 M2
#> truth 0.442 TRUE B T1 1.99 FALSE C T2
#> observed NA TRUE B T1 1.99 FALSE C T2
# we can threshold the data like this
threshold <- function(org_data, imputed){
# checks
stopifnot(NROW(org_data) == length(imputed),
is.list(imputed), is.data.frame(org_data))
# threshold
is_cont <- which(sapply(org_data, is.numeric))
is_bin <- which(sapply(org_data, is.logical))
is_ord <- which(sapply(org_data, is.ordered))
is_mult <- which(sapply(org_data, is.factor))
is_mult <- setdiff(is_mult, is_ord)
stopifnot(
length(is_cont) + length(is_bin) + length(is_ord) + length(is_mult) ==
NCOL(org_data))
is_cat <- c(is_bin, is_ord, is_mult)
trans_to_df <- function(x){
if(is.matrix(x))
as.data.frame(t(x))
else
as.data.frame( x )
}
out_cont <- trans_to_df(sapply(imputed, function(x) unlist(x[is_cont])))
out_cat <- trans_to_df(sapply(imputed, function(x)
sapply(x[is_cat], which.max)))
out <- cbind(out_cont, out_cat)
# set factor levels etc.
out <- out[, order(c(is_cont, is_bin, is_ord, is_mult))]
if(length(is_bin) > 0)
out[, is_bin] <- out[, is_bin] > 1L
if(length(is_ord) > 0)
for(i in is_ord)
out[[i]] <- ordered(
unlist(out[[i]]), labels = levels(org_data[, i]))
if(length(is_mult) > 0)
for(i in is_mult)
out[[i]] <- factor(
unlist(out[[i]]), labels = levels(org_data[, i]))
colnames(out) <- colnames(org_data)
out
}
thresh_dat <- threshold(dat$seen_obs, imp_res)
# compare thresholded data with observed and true data
head(thresh_dat)
#> C1 B1 O1 M1 C2 B2 O2 M2
#> 1 0.714 TRUE B T1 1.989 FALSE C T2
#> 2 0.206 FALSE A T2 0.745 TRUE B T3
#> 3 0.111 FALSE B T2 0.132 TRUE A T3
#> 4 0.846 FALSE C T2 1.137 TRUE D T3
#> 5 0.725 TRUE A T4 0.646 FALSE B T2
#> 6 0.320 FALSE C T3 0.492 FALSE A T2
head(dat$seen_obs) # observed data
#> C1 B1 O1 M1 C2 B2 O2 M2
#> 1 NA TRUE B T1 1.989 FALSE C T2
#> 2 0.206 NA A T2 NA TRUE B T3
#> 3 0.111 FALSE B T2 0.132 NA <NA> T3
#> 4 NA NA <NA> T2 1.137 TRUE <NA> T3
#> 5 NA TRUE <NA> T4 NA FALSE B <NA>
#> 6 0.320 FALSE <NA> T3 0.492 FALSE A T2
head(dat$truth_obs) # true data
#> C1 B1 O1 M1 C2 B2 O2 M2
#> 1 0.442 TRUE B T1 1.989 FALSE C T2
#> 2 0.206 FALSE A T2 0.639 TRUE B T3
#> 3 0.111 FALSE B T2 0.132 FALSE A T3
#> 4 2.645 FALSE B T2 1.137 TRUE B T3
#> 5 1.495 TRUE B T4 0.162 FALSE B T2
#> 6 0.320 FALSE C T3 0.492 FALSE A T2
# compare correct categories
get_classif_error <- function(impu_dat, truth = dat$truth_obs,
observed = dat$seen_obs){
is_cat <- sapply(truth, function(x)
is.logical(x) || is.factor(x))
is_match <- impu_dat[, is_cat] == truth[, is_cat]
is_match <- matrix(is_match, ncol = sum(is_cat))
is_match[!is.na(observed[, is_cat])] <- NA_integer_
setNames(1 - colMeans(is_match, na.rm = TRUE),
colnames(truth)[is_cat])
}
get_classif_error(thresh_dat)
#> B1 O1 M1 B2 O2 M2
#> 0.339 0.653 0.598 0.393 0.615 0.582
# compute RMSE
get_rmse <- function(impu_dat, truth = dat$truth_obs,
observed = dat$seen_obs){
is_con <- sapply(truth, is.numeric)
err <- as.matrix(impu_dat[, is_con] - truth[, is_con])
err[!is.na(observed[, is_con])] <- NA_real_
sqrt(colMeans(err^2, na.rm = TRUE))
}
get_rmse(thresh_dat)
#> C1 C2
#> 1.09 1.10
# we can compare this with missForest
miss_forest_arg <- dat$seen_obs
is_log <- sapply(miss_forest_arg, is.logical)
miss_forest_arg[, is_log] <- lapply(miss_forest_arg[, is_log], as.factor)
set.seed(1)
system.time(miss_res <- missForest(miss_forest_arg))
#> missForest iteration 1 in progress...done!
#> missForest iteration 2 in progress...done!
#> missForest iteration 3 in progress...done!
#> missForest iteration 4 in progress...done!
#> missForest iteration 5 in progress...done!
#> missForest iteration 6 in progress...done!
#> missForest iteration 7 in progress...done!
#> missForest iteration 8 in progress...done!
#> user system elapsed
#> 9.86 0.02 9.88
# turn binary variables back to logical variables
miss_res$ximp[, is_log] <- lapply(
miss_res$ximp[, is_log], function(x) as.integer(x) > 1L)
# compare errors
rbind(mdgc = get_classif_error(thresh_dat),
missForest = get_classif_error(miss_res$ximp))
#> B1 O1 M1 B2 O2 M2
#> mdgc 0.339 0.653 0.598 0.393 0.615 0.582
#> missForest 0.394 0.695 0.645 0.422 0.644 0.643
rbind(mdgc = get_rmse(thresh_dat),
missForest = get_rmse(miss_res$ximp))
#> C1 C2
#> mdgc 1.09 1.10
#> missForest 1.12 1.09We make a small example below were we take the iris data set and randomly mask it. Then we compare the imputation method in this package with missForest.
# re-scales continuous variables to have scale 1.
#
# Args:
# dat: data to rescale.
re_scale <- function(dat){
is_num <- sapply(dat, is.numeric)
if(!any(is_num))
return(dat)
dat[is_num] <- lapply(dat[is_num], scale)
dat[is_num] <- lapply(dat[is_num], c)
dat
}
# load the iris data set
data(iris)
iris <- re_scale(iris)
# assign function to produce iris data set with NAs.
#
# Args:
# dat: data set to mask.
# p_na: chance of missing a value.
mask_dat <- function(dat, p_na = .3){
is_miss <- matrix(p_na > runif(NROW(dat) * NCOL(dat)),
NROW(dat), NCOL(dat))
while(any(all_missing <- apply(is_miss, 1, all)))
# avoid rows with all missing variables
is_miss[all_missing, ] <- p_na > runif(sum(all_missing) * NCOL(dat))
# create data set with missing values
out <- dat
out[is_miss] <- NA
out
}
# get a data set with all missing values
set.seed(68129371)
dat <- mask_dat(iris)
# use the mdgc method
system.time(
mdgc_res <- mdgc(dat, maxpts = 10000L, minvls = 500L, n_threads = 4L,
maxit = 50L, use_aprx = TRUE, conv_crit = 1e-8,
method = "svrg", rel_eps = 1e-2, batch_size = 100L,
iminvls = 2000L, imaxit = 20000L, irel_eps = 1e-3,
lr = 1e-3))
#> user system elapsed
#> 2.052 0.000 0.523
# some of the imputed values
head(mdgc_res$ximp)
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> 1 -0.8977 1.0156 -1.34 -1.311 setosa
#> 2 -1.1392 -0.1315 -1.34 -1.311 setosa
#> 3 -1.3807 0.3273 -1.39 -1.311 setosa
#> 4 -1.5015 0.0979 -1.28 -1.311 setosa
#> 5 -0.5354 1.2450 -1.28 -1.180 setosa
#> 6 0.0427 1.9333 -1.17 0.125 setosa
# compare with missForest
system.time(miss_res <- missForest(dat))
#> missForest iteration 1 in progress...done!
#> missForest iteration 2 in progress...done!
#> missForest iteration 3 in progress...done!
#> missForest iteration 4 in progress...done!
#> missForest iteration 5 in progress...done!
#> missForest iteration 6 in progress...done!
#> missForest iteration 7 in progress...done!
#> missForest iteration 8 in progress...done!
#> user system elapsed
#> 0.445 0.012 0.457
# the errors
rbind(
mdgc = get_classif_error(
impu_dat = mdgc_res$ximp, truth = iris, observed = dat),
missForest = get_classif_error(
impu_dat = miss_res$ximp, truth = iris, observed = dat))
#> Species
#> mdgc 0.1905
#> missForest 0.0794
rbind(
mdgc = get_rmse(
impu_dat = mdgc_res$ximp, truth = iris, observed = dat),
missForest =
get_rmse(impu_dat = miss_res$ximp, truth = iris, observed = dat))
#> Sepal.Length Sepal.Width Petal.Length Petal.Width
#> mdgc 0.583 0.826 0.322 0.550
#> missForest 0.571 0.619 0.359 0.407We repeat this a few times to get Monte Carlo estimates of the errors:
# function to get Monte Carlo estimates of the errors.
#
# Args:
# dat: data set to use.
# seeds: seeds to use.
get_err_n_time <- function(dat, seeds){
cl <- makeCluster(4L)
registerDoParallel(cl)
on.exit(stopCluster(cl))
sapply(seeds, function(s){
# mask data
set.seed(s)
dat_mask <- mask_dat(dat)
# fit models
mdgc_time <- system.time(
mdgc_res <- mdgc(dat_mask, maxpts = 10000L, minvls = 500L, n_threads = 4L,
maxit = 50L, use_aprx = TRUE, conv_crit = 1e-8,
method = "svrg", rel_eps = 1e-2, batch_size = 100L,
iminvls = 2000L, imaxit = 20000L, irel_eps = 1e-3,
lr = 1e-3))
# compare with missForest
miss_forest_arg <- dat_mask
is_log <- sapply(miss_forest_arg, is.logical)
miss_forest_arg[, is_log] <- lapply(miss_forest_arg[, is_log], as.factor)
sink(tempfile())
miss_time <- system.time(miss_res <- missForest(
miss_forest_arg, parallelize = "forests"))
sink()
# turn binary variables back to logicals
miss_res$ximp[, is_log] <- lapply(
miss_res$ximp[, is_log], function(x) as.integer(x) > 1L)
# get the errors
er_int <- rbind(
mdgc = get_classif_error(
impu_dat = mdgc_res$ximp, truth = dat, observed = dat_mask),
missForest = get_classif_error(
impu_dat = miss_res$ximp, truth = dat, observed = dat_mask))
er_con <- rbind(
mdgc = get_rmse(
impu_dat = mdgc_res$ximp, truth = dat, observed = dat_mask),
missForest =
get_rmse(impu_dat = miss_res$ximp, truth = dat, observed = dat_mask))
# gather the output and return
er <- cbind(er_int, er_con)
er <- er[, match(colnames(er), colnames(dat))]
ti <- rbind(mdgc_time, miss_time)[, 1:3]
out <- cbind(er, ti)
message(sprintf("\nResult with seed %d is:", s))
message(paste0(capture.output(print(out)), collapse = "\n"))
out
}, simplify = "array")
}
# get the results
seeds <- c(40574428L, 13927943L, 31430660L, 38396447L, 20137114L, 59492953L,
93913797L, 95452931L, 77261969L, 10996196L)
res <- get_err_n_time(iris, seeds)# compute means and Monte Carlo standard errors
show_res <- function(res, dat){
stats <- apply(res, 1:2, function(x)
c(mean = mean(x), SE = sd(x) / sqrt(length(x))))
stats <- stats[, , c(colnames(dat), "user.self", "elapsed")]
for(i in seq_len(dim(stats)[[3]])){
nam <- dimnames(stats)[[3]][i]
cap <- if(nam %in% colnames(dat)){
if(is.ordered(dat[[nam]]))
"ordinal"
else if(is.factor(dat[[nam]]))
"multinomial"
else if(is.logical(dat[[nam]]))
"binary"
else
"continuous"
} else
"computation time"
cat(sprintf("\n%s (%s):\n", nam, cap))
print(apply(round(
stats[, , i], 4), 2, function(x) sprintf("%.4f (%.4f)", x[1], x[2])),
quote = FALSE)
}
}
show_res(res, iris)
#>
#> Sepal.Length (continuous):
#> mdgc missForest
#> 0.5547 (0.0128) 0.5130 (0.0268)
#>
#> Sepal.Width (continuous):
#> mdgc missForest
#> 0.8467 (0.0193) 0.7319 (0.0222)
#>
#> Petal.Length (continuous):
#> mdgc missForest
#> 0.2992 (0.0263) 0.2380 (0.0256)
#>
#> Petal.Width (continuous):
#> mdgc missForest
#> 0.4056 (0.0160) 0.3271 (0.0276)
#>
#> Species (multinomial):
#> mdgc missForest
#> 0.1664 (0.0140) 0.0875 (0.0092)
#>
#> user.self (computation time):
#> mdgc missForest
#> 1.7445 (0.2599) 1.1443 (0.0592)
#>
#> elapsed (computation time):
#> mdgc missForest
#> 0.4465 (0.0650) 4.1789 (0.2012)We do as in the Edgar Anderson’s Iris Data section here but with a different data set.
# prepare the data
library(survival)
colon_use <- colon[, setdiff(
colnames(colon), c("id", "study", "time", "status", "node4", "etype"))]
colon_use <- within(colon_use, {
sex <- sex > 0
obstruct <- obstruct > 0
perfor <- perfor > 0
adhere <- adhere > 0
differ <- ordered(differ)
extent <- ordered(extent)
surg <- surg > 0
})
colon_use <- colon_use[complete.cases(colon_use), ]
colon_use <- re_scale(colon_use)
# stats for the data set
summary(colon_use)
#> rx sex age obstruct perfor
#> Obs :610 Mode :logical Min. :-3.51 Mode :logical Mode :logical
#> Lev :588 FALSE:856 1st Qu.:-0.57 FALSE:1434 FALSE:1722
#> Lev+5FU:578 TRUE :920 Median : 0.10 TRUE :342 TRUE :54
#> Mean : 0.00
#> 3rd Qu.: 0.77
#> Max. : 2.11
#> adhere nodes differ extent surg
#> Mode :logical Min. :-1.04 1: 180 1: 38 Mode :logical
#> FALSE:1520 1st Qu.:-0.75 2:1306 2: 204 FALSE:1300
#> TRUE :256 Median :-0.47 3: 290 3:1460 TRUE :476
#> Mean : 0.00 4: 74
#> 3rd Qu.: 0.38
#> Max. : 8.29
# sample missing values
set.seed(68129371)
dat <- mask_dat(colon_use)
# use the mdgc method
system.time(
mdgc_res <- mdgc(dat, maxpts = 10000L, minvls = 500L, n_threads = 4L,
maxit = 50L, use_aprx = TRUE, conv_crit = 1e-8,
method = "svrg", rel_eps = 1e-2, batch_size = 100L,
iminvls = 2000L, imaxit = 20000L, irel_eps = 1e-3,
lr = 1e-3))
#> user system elapsed
#> 111.549 0.008 28.124
# some of the imputed values
head(mdgc_res$ximp)
#> rx sex age obstruct perfor adhere nodes differ extent surg
#> 1 Lev+5FU TRUE -1.4114 FALSE FALSE FALSE 0.378 2 3 FALSE
#> 2 Lev+5FU FALSE -1.4114 FALSE FALSE FALSE -0.187 2 3 FALSE
#> 3 Lev+5FU TRUE 0.0998 FALSE FALSE FALSE -0.753 2 3 FALSE
#> 4 Lev+5FU TRUE 0.0998 FALSE FALSE FALSE -0.753 2 3 FALSE
#> 5 Lev+5FU FALSE 0.9394 FALSE FALSE FALSE 0.943 2 3 FALSE
#> 6 Lev+5FU FALSE 0.3517 FALSE FALSE TRUE -0.470 2 2 FALSE
# compare with missForest
miss_forest_arg <- dat
is_log <- sapply(miss_forest_arg, is.logical)
miss_forest_arg[, is_log] <- lapply(miss_forest_arg[, is_log], as.factor)
system.time(miss_res <- missForest(miss_forest_arg))
#> missForest iteration 1 in progress...done!
#> missForest iteration 2 in progress...done!
#> missForest iteration 3 in progress...done!
#> missForest iteration 4 in progress...done!
#> missForest iteration 5 in progress...done!
#> missForest iteration 6 in progress...done!
#> missForest iteration 7 in progress...done!
#> missForest iteration 8 in progress...done!
#> user system elapsed
#> 9.856 0.008 9.864
# turn binary variables back to logicals
miss_res$ximp[, is_log] <- lapply(
miss_res$ximp[, is_log], function(x) as.integer(x) > 1L)
# the errors
rbind(
mdgc = get_classif_error(
impu_dat = mdgc_res$ximp, truth = colon_use, observed = dat),
missForest = get_classif_error(
impu_dat = miss_res$ximp, truth = colon_use, observed = dat))
#> rx sex obstruct perfor adhere differ extent surg
#> mdgc 0.641 0.520 0.191 0.0300 0.122 0.260 0.160 0.257
#> missForest 0.588 0.427 0.239 0.0229 0.218 0.439 0.327 0.337
rbind(
mdgc = get_rmse(
impu_dat = mdgc_res$ximp, truth = colon_use, observed = dat),
missForest = get_rmse(
impu_dat = miss_res$ximp, truth = colon_use, observed = dat))
#> age nodes
#> mdgc 1.02 1.11
#> missForest 1.10 1.08# get the results
res <- get_err_n_time(colon_use, seeds)# compute means and Monte Carlo standard errors
show_res(res, colon_use)
#>
#> rx (multinomial):
#> mdgc missForest
#> 0.6693 (0.0077) 0.6238 (0.0075)
#>
#> sex (binary):
#> mdgc missForest
#> 0.4784 (0.0073) 0.4374 (0.0072)
#>
#> age (continuous):
#> mdgc missForest
#> 1.0005 (0.0066) 1.0595 (0.0073)
#>
#> obstruct (binary):
#> mdgc missForest
#> 0.2059 (0.0044) 0.2858 (0.0047)
#>
#> perfor (binary):
#> mdgc missForest
#> 0.0297 (0.0017) 0.0445 (0.0029)
#>
#> adhere (binary):
#> mdgc missForest
#> 0.1508 (0.0038) 0.2326 (0.0071)
#>
#> nodes (continuous):
#> mdgc missForest
#> 1.0836 (0.0286) 1.0804 (0.0151)
#>
#> differ (ordinal):
#> mdgc missForest
#> 0.2636 (0.0037) 0.4562 (0.0097)
#>
#> extent (ordinal):
#> mdgc missForest
#> 0.1825 (0.0032) 0.3489 (0.0093)
#>
#> surg (binary):
#> mdgc missForest
#> 0.2650 (0.0041) 0.3589 (0.0077)
#>
#> user.self (computation time):
#> mdgc missForest
#> 113.6367 (7.6928) 3.0040 (0.1383)
#>
#> elapsed (computation time):
#> mdgc missForest
#> 28.6709 (1.9266) 11.7794 (0.4941)We do as in the Edgar Anderson’s Iris Data section here but with a different data set.
# prepare the data
data("nhanes", package = "survey")
nhanes_use <- within(nhanes, {
HI_CHOL <- HI_CHOL > 0
race <- factor(race)
agecat <- ordered(agecat)
RIAGENDR <- RIAGENDR > 1
})[, c("HI_CHOL", "race", "agecat", "RIAGENDR")]
nhanes_use <- nhanes_use[complete.cases(nhanes_use), ]
nhanes_use <- re_scale(nhanes_use)
# summary stats for the data
summary(nhanes_use)
#> HI_CHOL race agecat RIAGENDR
#> Mode :logical 1:2532 (0,19] :2150 Mode :logical
#> FALSE:7059 2:3450 (19,39] :1905 FALSE:3889
#> TRUE :787 3:1406 (39,59] :1911 TRUE :3957
#> 4: 458 (59,Inf]:1880
# sample a data set
set.seed(1)
dat <- mask_dat(nhanes_use)
# use the mdgc method
system.time(
mdgc_res <- mdgc(dat, maxpts = 10000L, minvls = 500L, n_threads = 4L,
maxit = 50L, use_aprx = TRUE, conv_crit = 1e-8,
method = "svrg", rel_eps = 1e-2, batch_size = 100L,
iminvls = 2000L, imaxit = 20000L, irel_eps = 1e-3,
lr = 1e-3))
#> user system elapsed
#> 58.1 0.0 14.9
# some of the imputed values
head(mdgc_res$ximp)
#> HI_CHOL race agecat RIAGENDR
#> 1 FALSE 2 (19,39] FALSE
#> 2 FALSE 3 (0,19] FALSE
#> 3 FALSE 3 (0,19] FALSE
#> 4 FALSE 3 (59,Inf] TRUE
#> 5 FALSE 1 (19,39] FALSE
#> 6 TRUE 2 (39,59] TRUE
# compare with missForest
miss_forest_arg <- dat
is_log <- sapply(miss_forest_arg, is.logical)
miss_forest_arg[, is_log] <- lapply(miss_forest_arg[, is_log], as.factor)
system.time(miss_res <- missForest(miss_forest_arg))
#> missForest iteration 1 in progress...done!
#> missForest iteration 2 in progress...done!
#> missForest iteration 3 in progress...done!
#> missForest iteration 4 in progress...done!
#> user system elapsed
#> 2.69 0.04 2.73
# turn binary variables back to logicals
miss_res$ximp[, is_log] <- lapply(
miss_res$ximp[, is_log], function(x) as.integer(x) > 1L)
# the errors
rbind(
mdgc = get_classif_error(
impu_dat = mdgc_res$ximp, truth = nhanes_use, observed = dat),
missForest = get_classif_error(
impu_dat = miss_res$ximp, truth = nhanes_use, observed = dat))
#> HI_CHOL race agecat RIAGENDR
#> mdgc 0.104 0.536 0.676 0.499
#> missForest 0.316 0.628 0.719 0.476# get the results
res <- get_err_n_time(nhanes_use, seeds)# compute means and Monte Carlo standard errors
show_res(res, nhanes_use)
#>
#> HI_CHOL (binary):
#> mdgc missForest
#> 0.0997 (0.0016) 0.3075 (0.0060)
#>
#> race (multinomial):
#> mdgc missForest
#> 0.5395 (0.0020) 0.7118 (0.0136)
#>
#> agecat (ordinal):
#> mdgc missForest
#> 0.6775 (0.0022) 0.6946 (0.0039)
#>
#> RIAGENDR (binary):
#> mdgc missForest
#> 0.4956 (0.0033) 0.4863 (0.0033)
#>
#> user.self (computation time):
#> mdgc missForest
#> 47.4060 (8.2625) 0.8179 (0.0686)
#>
#> elapsed (computation time):
#> mdgc missForest
#> 12.2692 (2.0692) 2.8193 (0.2150)Christoffersen, Benjamin, Mark Clements, Keith Humphreys, and Hedvig Kjellström. 2021. “Asymptotically Exact and Fast Gaussian Copula Models for Imputation of Mixed Data Types.” http://arxiv.org/abs/2102.02642.
Genz, Alan, and Frank Bretz. 2002. “Comparison of Methods for the Computation of Multivariate T Probabilities.” Journal of Computational and Graphical Statistics 11 (4): 950–71. https://doi.org/10.1198/106186002394.
Hoff, Peter D. 2007. “Extending the Rank Likelihood for Semiparametric Copula Estimation.” Ann. Appl. Stat. 1 (1): 265–83. https://doi.org/10.1214/07-AOAS107.
Zhao, Yuxuan, and Madeleine Udell. 2020a. “Matrix Completion with Quantified Uncertainty Through Low Rank Gaussian Copula.” In Advances in Neural Information Processing Systems (Neurips). http://arxiv.org/abs/2006.10829.
———. 2020b. “Missing Value Imputation for Mixed Data via Gaussian Copula.” In Proceedings of the 26th Acm Sigkdd International Conference on Knowledge Discovery & Data Mining, 636–46. KDD ’20. New York, NY, USA: Association for Computing Machinery. https://doi.org/10.1145/3394486.3403106.