The standard normal CDF
is an important function in a broad range of statistical
problems. When we need to evaluate the function many times (for example
in numerical integration), the computation performance may become an
issue.
One way to fast evaluate the function is to use a look-up table, that
is, we pre-compute a set of pairs ` and then use interpolation to approximate the
function value of a given
.
This simple library calculates the
function using piecewise linear interpolation. The
approximation error is guaranteed to be no greater than
.
The package can be installed from Github by calling:
remotes::install_github("boennecd/fastncdf")We need to first determine the knots
that we want to
pre-compute. Since
, we only need to consider non-negative
’s.
For , we set
and hence the error is bounded by
. Let
,
,
, where
is the smallest integer such that
. Then we need to determine the interval width
to satisfy the error
bound.
For piecewise linear interpolation, the error is bounded by
(Source http://pages.cs.wisc.edu/~amos/412/lecture-notes/lecture09.pdf)
Since , it can be shown that
.
Therefore, can be
calculated as:
(h <- sqrt(8 / dnorm(1) * 1e-7))## [1] 0.001818292
x <- seq(0, qnorm(1 - 1e-7) + h, by = h)
length(x)## [1] 2861
y <- pnorm(x)We can then call dput(x) and dput(y) to get the data we need.
We compare the speed of fastpnorm() and fastpnorm_preallocated()
with pnorm() in R:
library(fastncdf)
u <- seq(-6, 6, by = 1e-6)
system.time(truth <- pnorm(u))## user system elapsed
## 0.487 0.028 0.516
system.time(fasty <- fastpnorm(u))## user system elapsed
## 0.041 0.020 0.060
system.time(fasty_prec <- fastpnorm(u, TRUE))## user system elapsed
## 0.130 0.016 0.146
range(truth - fasty)## [1] -9.99999e-08 9.99999e-08
range(truth - fasty_prec)## [1] -9.99999e-08 9.99999e-08
# if we already had a vector with values then we can use a faster version
res <- rep(0., length(u))
system.time(fastpnorm_preallocated(u, res))## user system elapsed
## 0.03 0.00 0.03
all.equal(res, fasty)## [1] TRUE
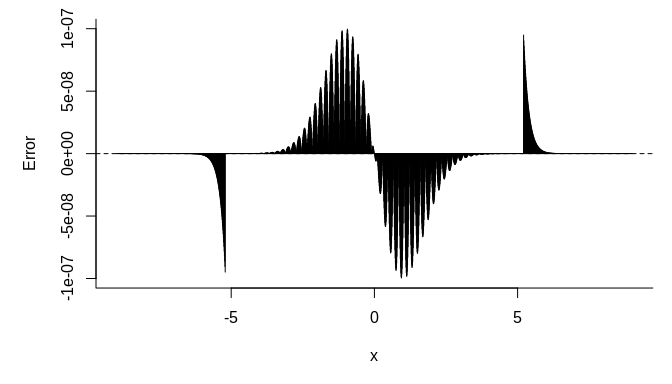
We plot the error versus the quantile below:
par(mar = c(5, 5, 1, 1))
us <- seq(-9, 9, length.out = 2000)
plot(us, fastpnorm(us) - pnorm(us), type = "h",
bty = "l", xlab = expression(x), ylab = "Error")
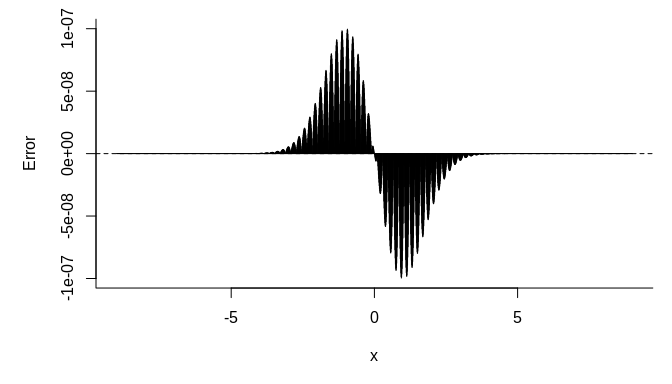
abline(h = 0, lty = 2)plot(us, fastpnorm(us, TRUE) - pnorm(us), type = "h",
bty = "l", xlab = expression(x), ylab = "Error")
abline(h = 0, lty = 2)We can get a similar result using R’s approxfun to do linear
interpolation:
lin_aprx <- local({
f <- approxfun(x = x, y = y, yleft = 0.5, yright = 1)
function(x){
p <- f(abs(x))
ifelse(x < 0, 1 - p, p)
}
})
max(abs(lin_aprx(u) - fastpnorm(u)))## [1] 1.554312e-15
The R version is slower though:
system.time(lin_aprx(u))## user system elapsed
## 0.475 0.156 0.631
system.time(fastpnorm(u))## user system elapsed
## 0.032 0.028 0.060
We can though use R’s splinefun to see the performance of other
functions. In particular, we can consider monotone cubic interpolation
using Fritsch–Carlson method:
m_splin <- local({
n_points <- 300L
eps <- 1e-9
x <- seq(0, qnorm(1 - eps), length.out = n_points)
f <- splinefun(x = x, y = pnorm(x), method = "monoH.FC")
x_max <- max(x)
function(x){
p <- f(abs(x))
out <- ifelse(x < 0, 1 - p, p)
ifelse(abs(x) > x_max, .5 * (1 + sign(x)), out)
}
})
# check the error
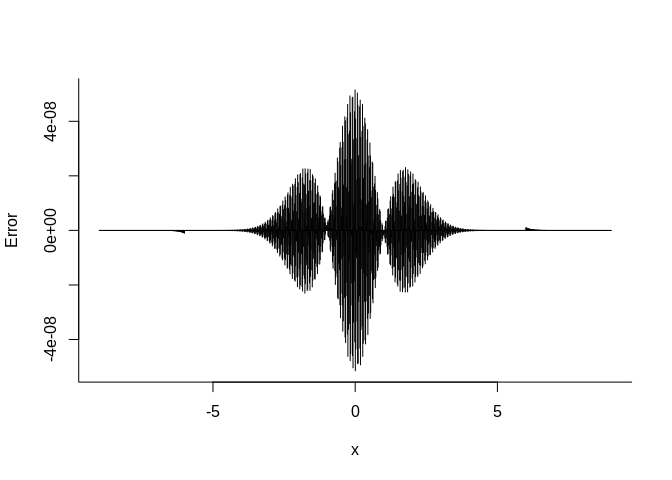
range(truth - m_splin(u))## [1] -5.165321e-08 5.165321e-08
# plot the error
plot(us, m_splin(us) - pnorm(us), type = "h",
bty = "l", xlab = expression(x), ylab = "Error")We will require three doubles per knot unlike the two we need for the linear interpolation. Furthermore, more computation is needed to perform the interpolation. However, we may need much fewer knots as shown above and this will reduce the cache misses.
A C++ implementation is also provided with this package:
all.equal(m_splin(u), fastpnorm(u, use_cubic = TRUE))## [1] TRUE
system.time(aprx_cubic <- fastpnorm(u, use_cubic = TRUE))## user system elapsed
## 0.076 0.024 0.101
max(abs(aprx_cubic - truth))## [1] 5.165321e-08
res <- rep(0., length(u))
system.time(fastpnorm_preallocated(u, res, use_cubic = TRUE))## user system elapsed
## 0.073 0.000 0.073
all.equal(res, aprx_cubic)## [1] TRUE
We can compare the monotone cubic interpolation with the linear interpolation with a particular focus on how well they scale in the number of threads used in the computation:
res <- rep(0, length(u))
test_func <- function(use_cubic, n_threads)
fastpnorm_preallocated(u, res, n_threads = n_threads,
use_cubic = use_cubic)
bench::mark(
`pnorm ` = pnorm(u),
`linear (1 thread) ` = test_func(FALSE, 1L),
`linear (2 threads)` = test_func(FALSE, 2L),
`linear (4 threads)` = test_func(FALSE, 4L),
`linear (6 threads)` = test_func(FALSE, 6L),
`cubic (1 thread) ` = test_func(TRUE , 1L),
`cubic (2 threads)` = test_func(TRUE , 2L),
`cubic (4 threads)` = test_func(TRUE , 4L),
`cubic (6 threads)` = test_func(TRUE , 6L),
check = FALSE, min_time = 2)## # A tibble: 9 x 6
## expression min median `itr/sec` mem_alloc `gc/sec`
## <bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>
## 1 pnorm 508.7ms 519.2ms 1.92 91.6MB 0
## 2 linear (1 thread) 29.2ms 30.3ms 32.8 0B 0
## 3 linear (2 threads) 18ms 19ms 51.7 668.5KB 0
## 4 linear (4 threads) 17.7ms 19ms 52.1 0B 0
## 5 linear (6 threads) 17.9ms 18.9ms 51.6 0B 0
## 6 cubic (1 thread) 68.9ms 69.6ms 14.2 0B 0
## 7 cubic (2 threads) 35.3ms 35.7ms 27.5 0B 0
## 8 cubic (4 threads) 18.7ms 20.2ms 49.0 0B 0
## 9 cubic (6 threads) 18.2ms 19ms 52.0 0B 0
We may prefer the monotone cubic interpolation given the lower error, lower memory requirements, it scales better in the number of threads, and it has less “sided” errors.