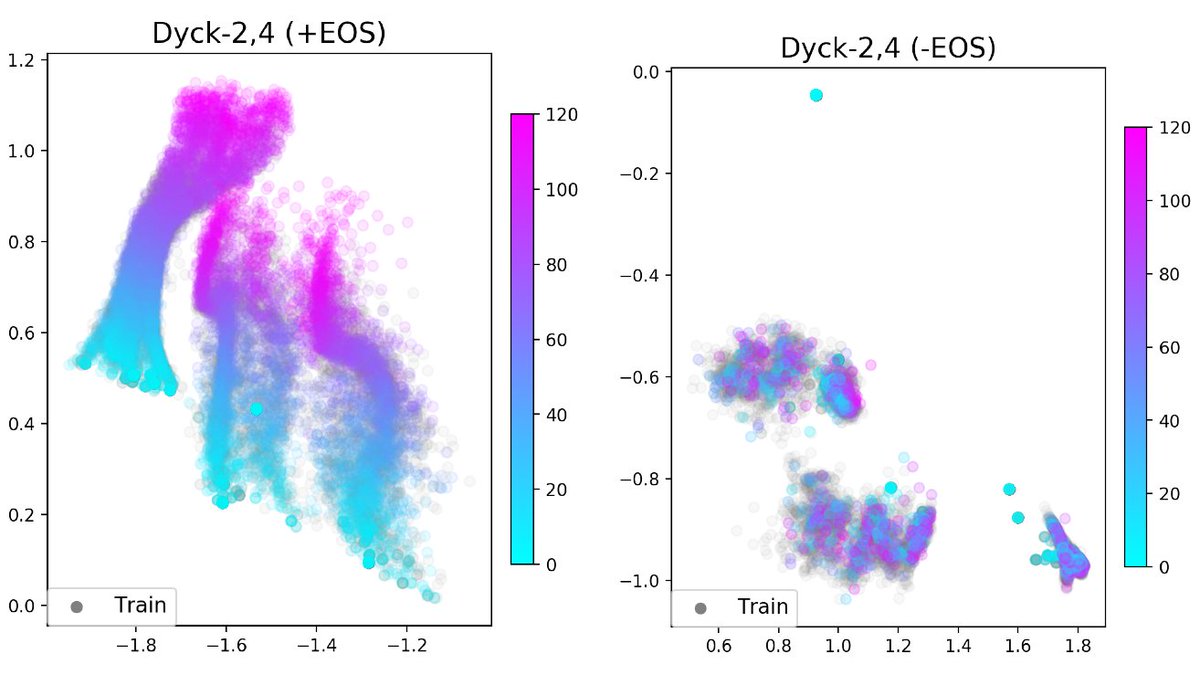
Current neural models are unable to generalize to sequences longer than those seen at training time, which is a barrier to achieving human-like language understanding. This problem is compounded by the widespread practice of using an end-of-sequence ([EOS]) token to mark the end of a training sequence. When these tokens are removed, models show a surprising ability to generalize to longer sequences in a number of domains. This repository contains experiments for investigating this phenomenon in three different settings.
The publication is available here.
- Clone the repository
git clone https://github.com/bnewm0609/eos-decision/
cd eos-decision- Install required packages
pip install -r requirements.txt- (Optional) Transformer Experiments To run the Transformer experiments, install the slightly modified version of the OpenNMT framework which is included as a git submodule of this repo. To install the modified version of the package, run:
cd OpenNMT-py
python setup.py install- configs: Stores the configuration files for the experiments. The fields of these files are detailed below.
- data: Stores the data from the tasks. All generated data is put in a subfolder in this directory.
- models: Is created when models are trained to store model weights.
- results: Is created when models are trained to store experiment results.
- src: Stores the code for running an experiment from a config file.
- scripts: Stores an assortment of scripts for running groups of experiments, generating and preprocessing data and generating results.
- tools/shared: Tokenization tools for the MT experiments written by by Philipp Koehn and Josh Schroeder. (Available at http://www.statmt.org/europarl/v7/tools.tgz)
To generate sequences, run:
sh scripts/dyck_km/generate_data.shTo train all of the models, use the run_all script:
python scripts/shared/run_all.py configs/dyck_km/all_experiments/ train-seq2seqThis will train all of the -EOS and +EOS models, with 5 random seeds of each. Use the --white_list flag to train only models whose configs contain a substring and the --black_list flag to train only models whose configs do not contain a substring.
To collect the mean close-bracket accuracy for an experiment, run the script here:
python scripts/dyck_km/collect_results.py configs/dyck_km/all_experiments/dyck{m}2-{eos,no_eos}/To gather perplexity, the parameter --metric ppl can be added.
To generate the plots seen in the paper run:
python scripts/dyck_km/generate_plots.py --save_hidden_statesTo train the probes to predict where sequences end, we can use the run_all script:
python scripts/shared/run_all.py configs/dyck_km/all_probes/ train-truncationThere are probes for +EOS and -EOS models separately for each of the 3 values of m, and 5 random seeds, giving 30 probes total.
The scripts/dyck_km/collect_results.py script can be used to collect these results as well with the --metric probe_acc flag.
Unlike the Dyck-(k,m) data, the SCAN data is already included in the repo.
To run the LSTM experiments, run:
python scripts/shared/run_all.py configs/scan/lstm_experiments train-seq2seqAs long as the OpenNMT submodule is installed, the transformer experiments are almost as easy. First, train the -EOS and +EOS models:
python scripts/shared/run_all.py configs/scan/transformer_experiments train-onmt --black_list "+eos_des+"This command also generates the outputs for the +EOS model and the -EOS+Oracle model.
To generate the output for the +EOS+Oracle model, run this command:
python scripts/shared/run_all.py configs/scan/transformer_experiments generate-onmt --white_list "+eos_des+"At this point, the transformer-generated outputs are saved to disk, so we have to run the evaluations for them. We can run the oracle length evaluation with the BLEU metric with the command:
python scripts/shared/run_all.py configs/scan/transformer_experiments evalTo generate the SCAN result tables in the paper, run:
python scripts/scan/collect_results.py configs/scan/{lstm,transformer}_experimentsTo get the lstm or transformer results respectively.
To generate the plots, run:
python scripts/dyck_km/generate_plots.py --save_hidden_statesTo download and preprocess the data, run
sh scripts/translation/generate_data.shThis will download the Europarl German-English training set from http://www.statmt.org/europarl/v9/training/, sample 500,000 examples, and partition it into a the length splits with length cutoffs of 10, 15, and 25 tokens.
Both the Transformer and LSTM experiments are conducted using OpenNMT, so there are three commands to run them all.
First, to train the -EOS and +EOS models and capture their generations:
python scripts/shared/run_all.py configs/translation/{lstm,transformer}_experiments train-onmt --black_list "+eos_des+"Next, to capture the generated sequences from the +EOS+Oracle models:
python scripts/shared/run_all.py configs/translation/{lstm,transformer}_experiments generate-onmt --white_list "+eos_des+"Finally, to evaluate using our the Oracle BLEU metric, run:
python scripts/shared/run_all.py configs/translation/{lstm,transformer}_experiments evalWe are using a slightly modified version of OpenNMT framework, which is included as a git submodule of this repo. To install the modified version of the package, run:
cd OpenNMT-py
python setup.py installSee the OpenNMT repository for more information.
The difference between the original repo and ours is just that we add a -disable_eos_sampling to the preprocess and translate scripts. In preprocess, this option replaces the EOS token with a padding token, and in the translate script, it prevents the model from predicting EOS tokens by setting their probability to 0.
Note: this is different from the custom code, where the use_eos options specifies what the data looks like and the disable_eos_sampling specifies what the decoder should do. To prevent naming conflicts, in the script for running OpenNMT experiments, we use the lm.use_eos entry of the config to decide whether to pass disable_eos_samplign to the preprocess script and the lm.disable_eos_sampling entry to decide whether to pass disable_eos_sampling to the translate script. This modification leads to behavior consistent with the custom experiments.
For information about preprocessing, training, and translating, see the OpenNMT documentation for more information.
For evaluations, we load the OpenNMT model outputs into a lookup-table-based model so we can evaluate them using the same methods as our custom-trained models. The lm_type in the lm section of the config specifies this lookup-table-based model.
Here is the overall structure of an experiment config:
First comes the information about the dataset:
data:
dataset_type: [selects the src.data.Dataset class to load in]
dataset_name: [labels in the model and results dirs, used for different splits]
train_path: [path to training data]
dev_path: [path to dev data]
test_path: [path to test data]
input_vocab_path: [path to preprocessed source vocab json]
target_vocab_path: [path to preprocess target vocab json]The dataset_type will be either scan, mt, dyckmk, or dyckkm_ending .
For Dyck-(k, m) experiments, the data section of config should look like:
data:
dataset_type: dyckmk # or dyckkm_ending for the EOS tokens
dataset_name: dyckmk{m}2 # m is the
train_path: [path to training data]/small_train.txt
dev_path: [path to dev data]/dev.txt
test_path: [path to test data]/tasks_test_length.txtFor SCAN experiments, the data section of the config should look similar:
data:
dataset_type: scan
dataset_name: scan{k}ls # this can be anything, but should be descriptive, where k is the length cutoff
train_path: [path to training data]/small_train.txt
dev_path: [path to dev data]/dev.txt
test_path: [path to test data]/tasks_test_length.txtThe vocabulary paths can point to wherever, but probably somewhere within the scan/length_{K}_split data directory.
For the MT experiments, the data section of the config should include:
data:
dataset_type: mt
dataset_name: mt_deennp500000ls{k} # this can be whatever, but should be descriptive and short
train_path: path/to/training.data
dev_path: path/to/dev.data
test_path: path/to/test.data
source_lang: de # two letter code, usually de
target_lang: en # usually enNext, comes information about the model, both the architecture and the training information:
lm:
lm_type: [selects the src.models model class to load in]
lm_path: [base path for saving the model]
[Architecture-specific information]
[Training-specific information]
batch_size: [int: batch size for models at train/dev/test-time]
use_eos: [bool: whether eos token are used in model training]
disable_eos_sampling: [bool: whether eos tokens are prohibited from being produced at test time]
seed: [int: random seed used for initialization and tracking multiple experiments]The lm_type is what indicates the type of model should be trained/evaluated. For LSTMs, it is seq2seq. For OpenNMT models, it should be s2s_lookup.
If use_eos is True, then we remove any [EOS] tokens from the target sequences when constructing the datasets (train, dev, and test).
If disable_eos_sampling is True, then at test time, the probability of the [EOS] token is set to 0 at each decoding step. (This only is used when use_eos is True.)
Now, the architecture-specific information looks different if the model is an LSTM or a transformer. For LSTMs, it includes information like:
encoder_cell_type: [str: 'standard' (LSTM) or 'GRU']
encoder_num_layers: [int: number of decoder layers]
decoder_cell_type: [str: 'standard' (LSTM) or 'GRU']
decoder_num_layers: [int: number of decoder layers]
embed_dim: [int: embedding dimension]
hidden_dim: [int: embedding dimension for both encoder and decoder]
dropout: [float: dropout probability between LSTM layers]An lm_type of s2s_lookup creates a basic look-up table whose keys are the input sequences in the data.dev_path and data.test_path.
lm_type: s2s_lookup
dev_output_path: "results/onmt_results/de_en50ls10hs500layers2Attn_npunct_des/dev.txt"
test_output_path: "results/onmt_results/de_en50ls10hs500layers2Attn_npunct_des/long.txt"
batch_size: 1This is used with OpenNMT experiments where model output is dumped to files using OpenNMT translation code, but we want to perform our own evaluations on it.
And finally, the training information includes:
batch_size: [int: size of training/dev/test batch]
lr: [float: learning rate. Usually 0.001 b/c we use Adam]
teacher_forcing_ratio: [float: percentage of examples where model output is fed into model
during training. Only used for LSTMs and usually is 0.5]Next, the truncation model, which predicts where the sequence should end
truncation:
model_path: [base path to where the truncation model is saved]
model_type: [indicates which truncation_model.TruncationModel is loaded in]
[Model-specific information]
lr: [learning rate]There are only two truncation models that are used: an oracle model that ends sequences at the optimal location and dyckkm_ending which trains the linear probe to predict where Dyck-(k,m) sequences can end.
For ONMT Transformers and LSTMs, we add another section labeled onmt which holds all of the parameters for the OpenNMT scripts. A minimal example might look like:
onmt:
preprocessing:
data_dir: "data/translation/de_en/subsampled_50/length_{k}_split/"
train_src: "small_train_de.txt"
train_tgt: "small_train_en.txt"
dev_src: "dev_de.txt"
dev_tgt: "dev_en.txt"
training:
save_model: "models/onmt_models/model_dir/"
world_size: 1
gpu_ranks: 0
translate:
model: "_step_10000.pt"
dev_src: "dev_de.txt"
test_src: "tasks_test_length_de.txt"
gpu: 0These arguments are a bit different from those in the OpenNMT pytorch docs. Here are the differences:
- Preprocessing:
dev_srcanddev_tgtreplacevalid_srcandvalid_tgt.train_src,train_tgt,dev_src, anddev_tgthave only the base name of the file, not the whole paths.data_dirreplacessave_data(and also provides the directory for the{train, dev}_{src, tgt}) files.
- Training:
datais replaced by preprocessing'sdata_dir.
- Translate:
modelhas the base name of the model we want to use, not the whole path. The directory is obtained from training'ssave_model.srcis replaced bydev_srcandtest_srcwhich hold the base names for the data files. The directory is obtained from preprocessing'sdata_dir.
View the OpenNMT pytorch docs for additional arguments to add (and descriptions of the ones we include in the config).
Finally, we have the reporter section which specifies which evaluations metrics to calculate.
reporter:
results_path: [path to directory where results are saved]
methods:
- [str: method one]
- [str: method two]
...Here are the possible evaluation metrics that can be listed in the methods category:
bleu: calculates the BLEU score of the predictions using the target as a reference using thesacrebleupackage (used for MT).close_bracket: calculates the bracket closing accuracy for the Dyck-(k,m) languages.dyck_ending: calculates the accuracy of the linear probe at predicting when Dyck-(k,m) sequences can end.exact_match: calculates proportion of the predictions that exactly match the targets (used for SCAN).perplexity: calculates the perplexity of the seq2seq model on the evaluation sets (used for SCAN and MT).oracle_eos: calculates the proportion of the predictions that exactly match the targets when the prediction is truncated to the true target length (used for SCAN).oracle_bleu: reports the bleu score between the predictions and targets where the prediction is truncated at the point that maximizes BLEU score. (used for MT but doesn't seem to work).seq2seq_samples: saves the model-generated sentences to disk.
If there are any other fields in the yaml file, they are not needed. You can see examples of configs in the configs directory.
@inproceedings{newman2020extrapolation,
author = {Benjamin Newman and John Hewitt and Percy Liang and Christopher D. Manning},
booktitle = {BlackBoxNLP@EMNLP},
link = {https://nlp.stanford.edu/pubs/newman2020extrapolation.pdf},
title = {The EOS Decision and Length Extrapolation},
url = {https://nlp.stanford.edu/pubs/newman2020extrapolation.pdf},
year = {2020}
}