CRISOT: Genome-wide CRISPR off-target prediction and optimization based on RNA-DNA interaction fingerprints
Read CRISOT paper via https://www.nature.com/articles/s41467-023-42695-4
✨The web server of CRISOT is now available at CRISOT web server (https://crisot.aigene.org.cn/).
🚗The CRISOT suite is also available via zenodo.8422214.
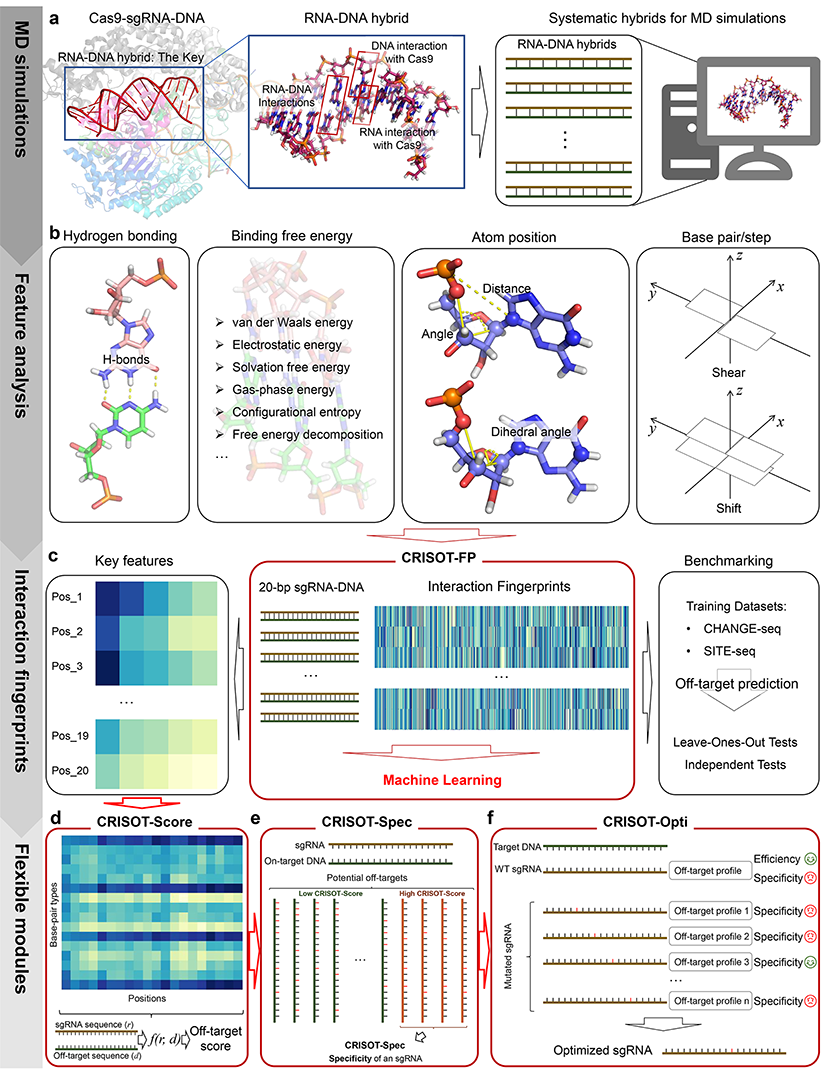
The powerful CRISPR-Cas9 genome editing system is hindered by its off-target effects. Although in silico tools have been developed for single-strand guide RNA (sgRNA) design and CRISPR off-target evaluation, the genome-wide CRISPR off-target prediction performance is limited. In this study, we proposed that the modelling of molecular interactions within the Cas9-sgRNA-DNA complex can substantially improve the genome-wide off-target prediction. To this end, we systematically performed molecular dynamics (MD) simulations to capture the molecular interaction features of the CRISPR-Cas9 system. CRISOT, which is a powerful tool package suite that contains various modules for genome-wide CRISPR off-target evaluation and sgRNA optimization, is presented.
In the CRISOT suite, CRISOT-FP was designed as the fingerprint of an sgRNA-DNA pair encoded by molecular interaction features, and an XGBoost (XGB) model trained with CRISOT-FP surpassed the state-of-the-art off-target prediction methods. Furthermore, CRISOT-Score was designed by using key interaction features identified from CRISOT-FP and has obtained great results in scoring off-target sites and distinguishing active off-targets from inactive ones. Then, CRISOT-Spec was designed to intuitively evaluate the off-target specificity of an sgRNA. Finally, CRISOT-Opti was designed for sgRNA optimization by ingeniously introducing a mutation to the sgRNA to reduce the off-target effect while maintaining efficient on-target effect.
- CRISOT-FP: Molecular interaction fingerprint for developing prediction models for CRISPR off-target effects. Users can use the trained models in models to predict CRISPR-Cas9 off-target effects.
- CRISOT-Score: Key interaction features were derived to develop CRISOT-Score, an efficient off-target scoring function to evaluate off-target effects.
- CRISOT-Spec: CRISOT-Spec is used to evaluate the off-target specificity of a given sgRNA.
- CRISOT-Opti: CRISOT-Opti is used for sgRNA optimization, which smartly introduces a single nucleotide mutation for the given sgRNA by reducing the off-target effect of the given sgRNA while maintaining its on-target effect.
Collectively, the CRISOT suite provides an accurate and comprehensive evaluation of genome-wide CRISPR off-target effects, as well as an efficient way to optimize a given sgRNA for improved off-target specificity in CRISPR genome editing.
- Python3
- xgboost==1.7.3
- pandas
- numpy
- Reference genome file (Can be downloaded from the UCSC website, hg38 is recommended)
- Cas-OFFinder==2.4
- GPU acceleration (Recommended)
- Create a python3 environment
- Install required python libraries
- Download and install Cas-OFFinder. NOTE: Properly set the environment variables to make Cas-OFFinder available.
- Download genome reference file from the UCSC website and uncompress it to data/. (e.g., data/hg38.fa).
- Unzip the trained models to models/ (Required only when using the CRISOT-FP prediction)
CRISOT suite includes several methods for off-target prediction and sgRNA optimization:
- score: CRISOT-Score of sgRNA-DNA, requires [--sgr, --tar];
- scores: Batch calculation of CRISOT-Score, requires [--csv], options [--on_item, --off_item, --out];
- spec: Calculate CRISOT-Spec, on-target sequence must be in the first line, requires [--csv], options [--on_item, --off_item];
- off_spec: Perform Cas-Offinder search and calculate CRISOT-Spec, requires [--sgr, --tar, --genome], options [--mm, --dev];
- rescore: Rescoring sgRNAs by CRISOT-Score and CRISOT-Spec, requires [--txt, --genome], options [--mm, --dev, --out];
- opti: CRISOT-Opti optimization by mutation, requires [--tar, --genome], options [--threshold, --mm, --dev, --out];
- crisot_fp: CRISOT-FP XGBoost machine learning prediction, requires [--csv], options [--xgb_model, --out, --on_item, --off_item]
- gen_fp: Generates the fingerprints for a given sgRNA-DNA pair, requires [--sgr, --tar], options [--out]
Calculates the CRISOT-Score score of a given pair of sgRNA-DNA, e.g.,
python CRISOT.py score --sgr GAGTCCGAGCAGAAGAAGAANGG --tar GAGTCCGAGCAGAAGAAGAANGG
# output:
CRISOT-Score:
0.9473928688501404python CRISOT.py score --sgr GAGTCCGAGCAGAAGAAGAANGG --tar GAGTCCGAGGAGAAGACGAAGGG
# output:
CRISOT-Score:
0.6892195294509914
NOTE: sgRNA and target sequences should be 23-nt, with NGG PAM.
Calculates CRISOT-Score scores of a series of sgRNA-DNA sequences in a csv file, e.g.,
python CRISOT.py scores --csv example/example.csv --out example/CRISOT-Score_example_out.csv
# output CRISOT-Score scores in file example/CRISOT-Score_example_out.csv
NOTE: The default column names of the input csv file are 'On' and 'Off' for sgRNA and target DNA sequences, respectively.
Given a series of off-target sequences of a specific sgRNA, spec calculates the CRISOT-Spec score, e.g.,
python CRISOT.py spec --csv example/example.csv
# output:
CRISOT-Spec:
0.8296024039222938
NOTE: Off-target sequences in the csv file should be of the same sgRNA. On-target sequence must be in the first line.
Given an sgRNA and its on-target sequence, off_spec searches for the genome-wide off-target sites and calculates the CRISOT-Spec score, e.g.,
python CRISOT.py off_spec --sgr GAGTCCGAGCAGAAGAAGAANGG --tar GAGTCCGAGCAGAAGAAGAAGGG --genome data/test_genome.fa
# output:
CRISOT-Spec:
0.912578202248319
NOTE: 1. Use the proper genome reference file. 2. Change the --dev if CUDA error occurs.
Rescoring and reranking the designed sgRNAs using the CRISOT-Score and CRISOT-Spec, e.g.,
python CRISOT.py rescore --txt example/example_sgRNAs.txt --genome data/test_genome.fa --out example/CRISOT_rescore_example_out.csv
# output rescoring results in file example/CRISOT_rescore_example_out.csv
NOTE: The input tsv file should contain sgRNA sequences in the 'Target sequence' column.
Optimizes a given sgRNA by smartly mutating the sgRNA, e.g.,
python CRISOT.py opti --tar GAGTCCGAGGAGAAGACGAAGGG --genome data/test_genome.fa --out example/CRISOT-Opti_example_out.csv
# output optimization results: example/CRISOT-Opti_example_out.csv
NOTE: The default CRISOT-Score threshold for the mutated sgRNAs is 0.8.
Evaluates the off-target effects using the XGBoost models trained using the CRISOT-FP, e.g.,
python CRISOT.py crisot_fp --csv example/example.csv --out example/CRISOT-FP_example_out.csv
# output CRISOT-FP results: example/CRISOT-FP_example_out.csv
NOTE & IMPORTANT: The trained XGBoost models are in models, please UNZIP them before using the crisot_fp. The training and testing datasets used in this study are available via zenodo.8420032.
Generates the CRISOT-FP encoding results of a given pair of sgRNA-DNA, e.g.,
python CRISOT.py gen_fp --sgr GAGTCCGAGCAGAAGAAGAANGG --tar GAGTCCGAGCAGAAGAAGAANGG --out example/CRISOT-FP_example_encoding.csv
# output CRISOT-FP encoding features: example/CRISOT-FP_example_encoding.csv
The CRISOT suite is licensed under the creative commons license CC BY-NC2.0 (https://creativecommons.org/licenses/by-nc/2.0/). The authors have filed a patent application on CRISOT. The paper is published on Nature Communications.