shinySeqBrowser is similar to genome browser tools (such as IGV), mainly built around the R package Gviz. I created this project to learn how to build R shiny apps and how to use the Gviz/GenomicFeatures/etc packages, all while providing something potentially useful for my lab. The main purpose is to be able to explore RNA-seq data across specific transcripts and easily export these plots for publications. This tool is still under development and feedback/issue reporting is welcome.
- Analyze read coverage of your BAM files
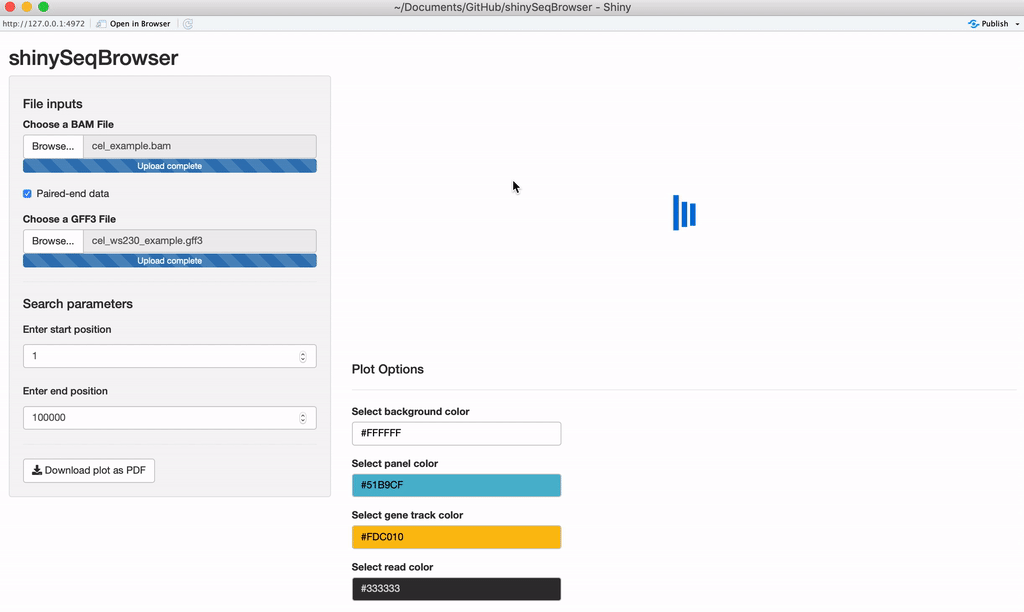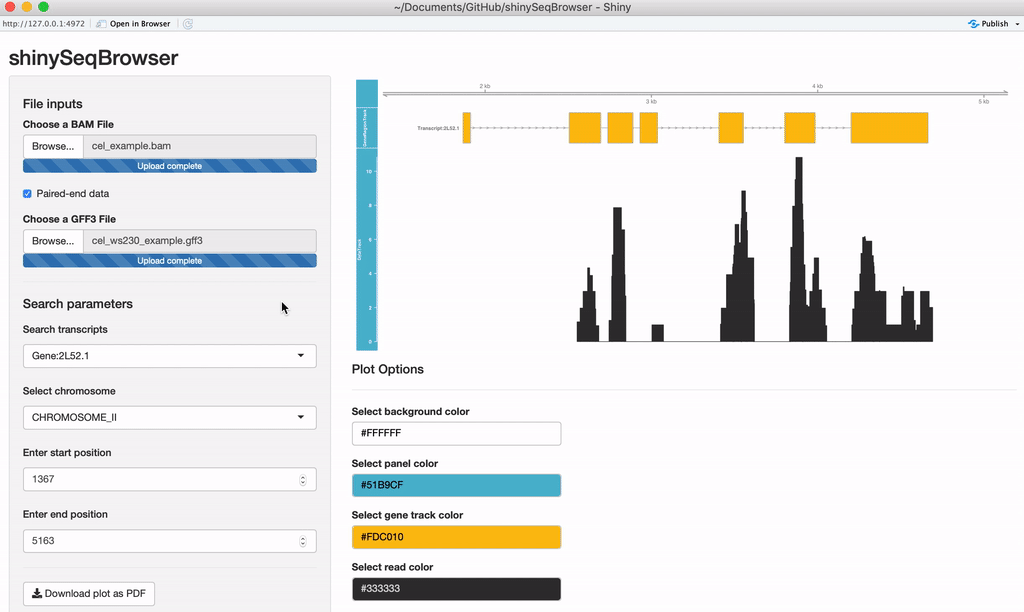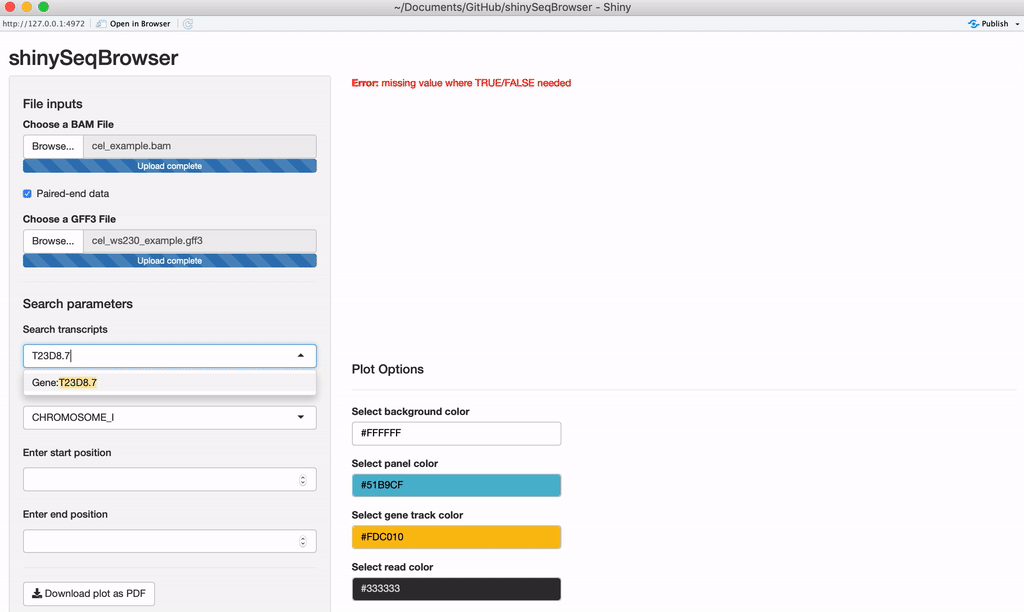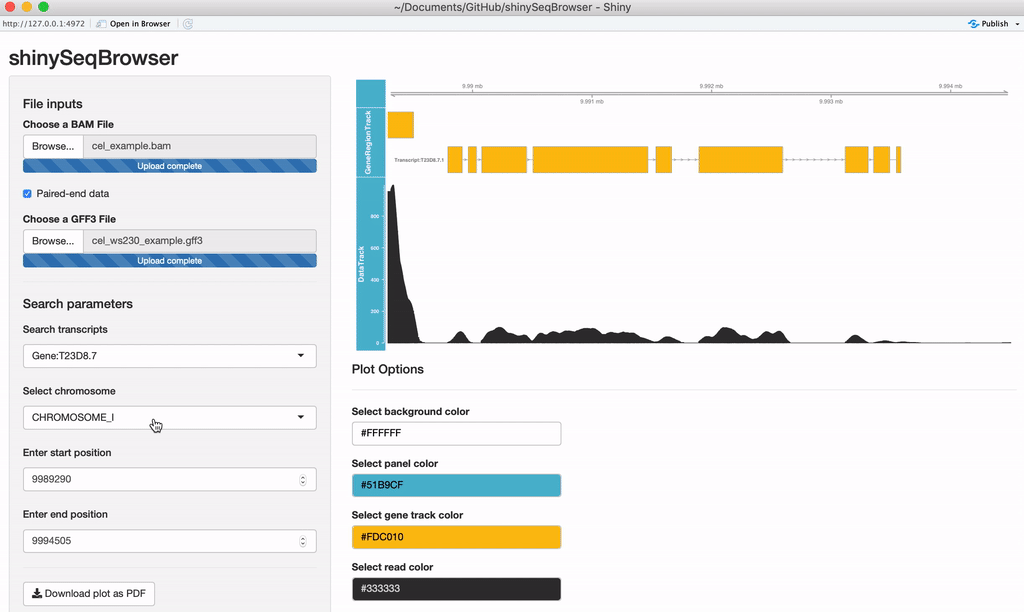
- Search transcripts by name, select regions by position/chromosome
- Customize styling and export plots for publication
In order to use this app you need to first install R from cran-r. I recommend that you also install RStudio to make your life easier if you plan on using R long-term.
Install the following dependencies from within R:
# Shiny related
install.packages("shiny")
install.packages("shinycssloader") # for the loader animation
install.packages("colourpicker") # for modifying plot colors
# Other packages
install.packages("Cairo") # for exporting PDF
# Bioconductor tools
source("http://bioconductor.org/biocLite.R")
biocLite("Gviz")
biocLite("Rsamtools")
biocLite("GenomicFeatures")
biocLite("GenomicRanges")
Then the easiest way to run the app is to run these commands from within R/RStudio:
library(shiny)
runGitHub("shinySeqBrowser", "biokcb")
If you want to clone the repository and make modifications to run on your local machine, you can use:
git clone https://github.com/biokcb/shinySeqBrowser.git
Then in R/Rstudio, run the app by directing the command to where the folder was downloaded after modifications.
library(shiny)
runApp('path/to/shinySeqBrowser/')
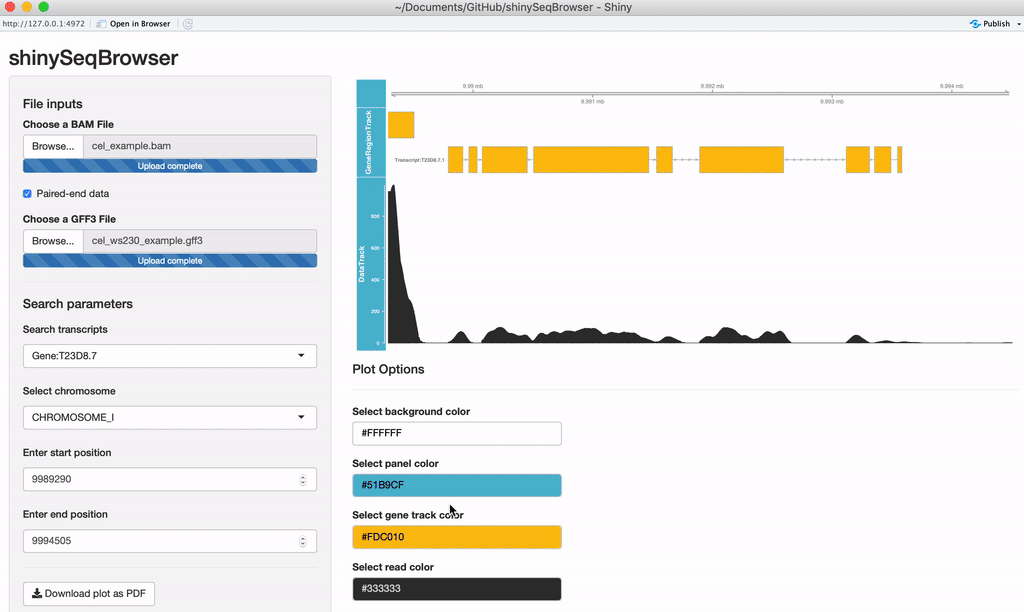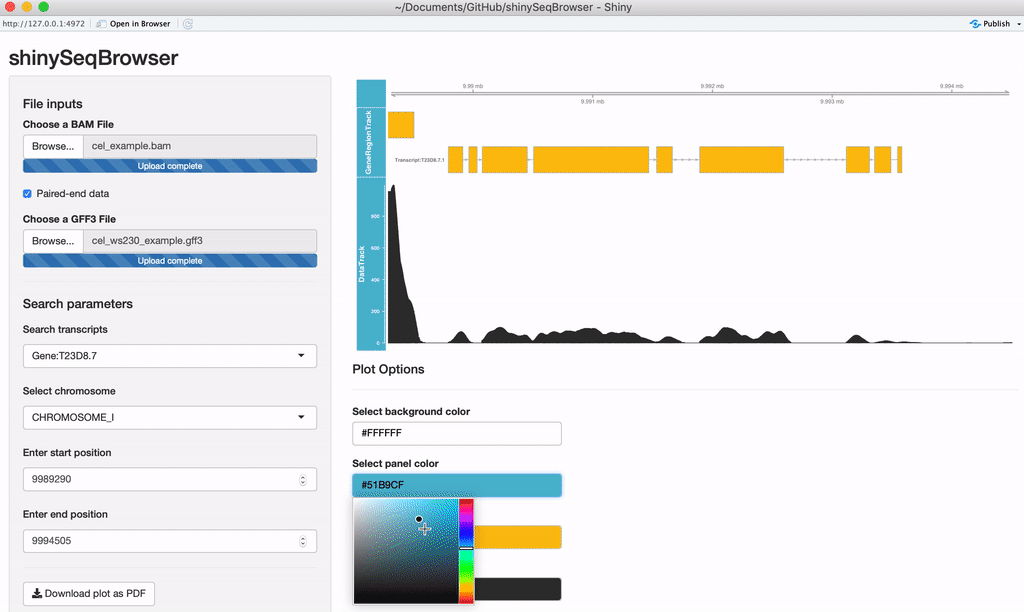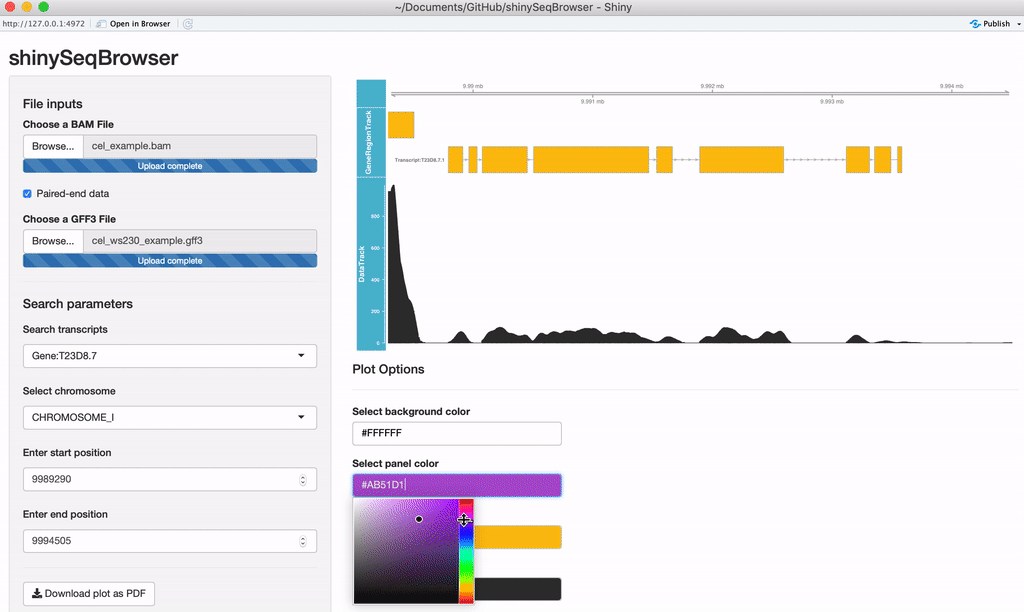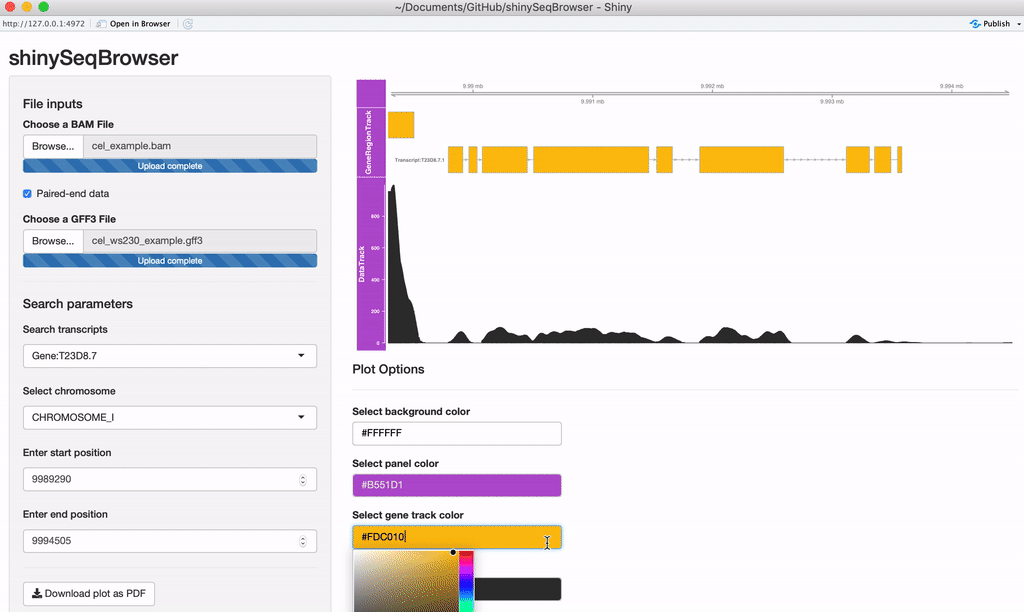
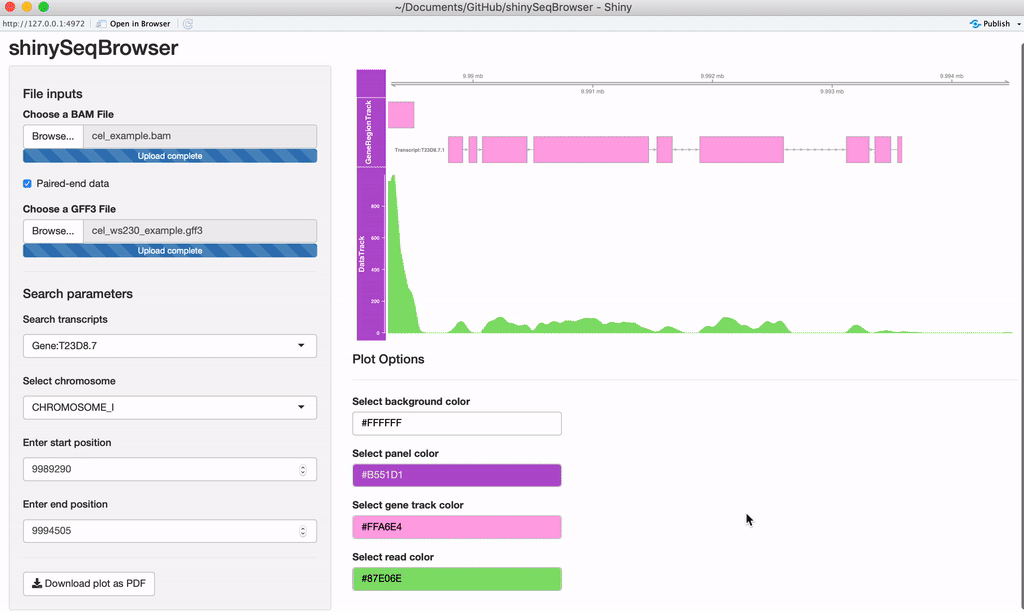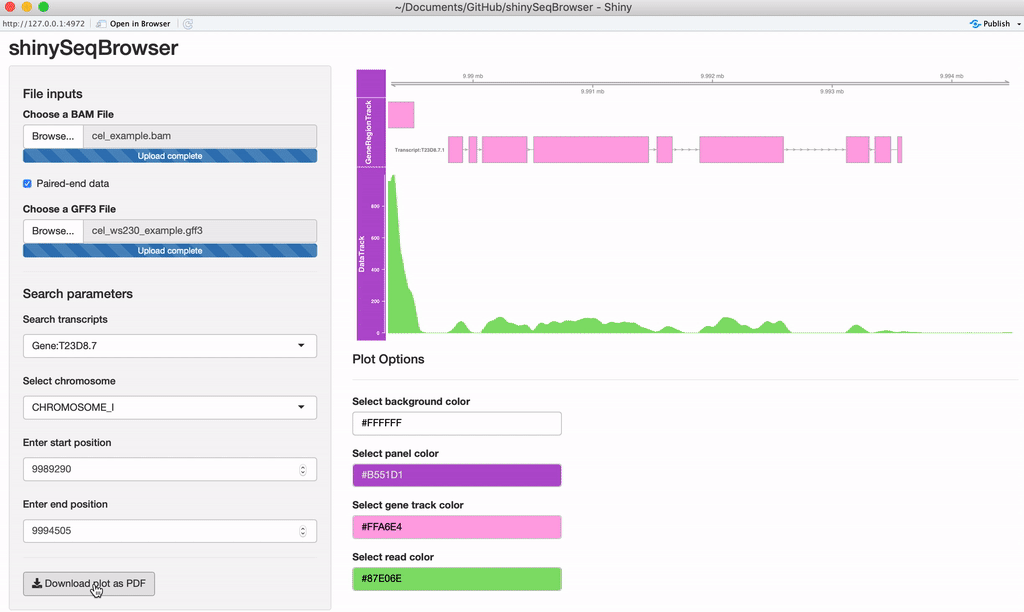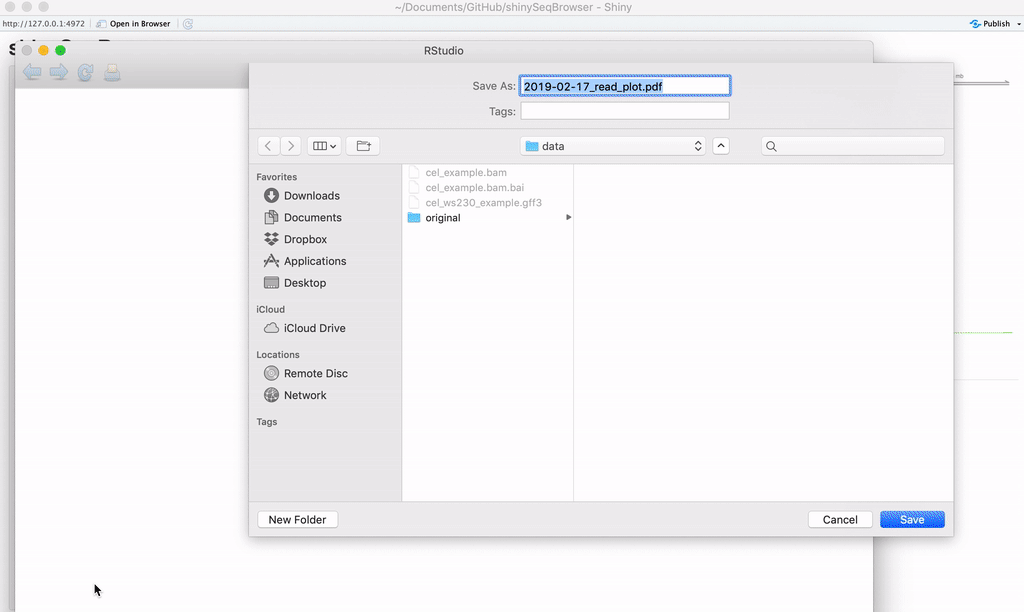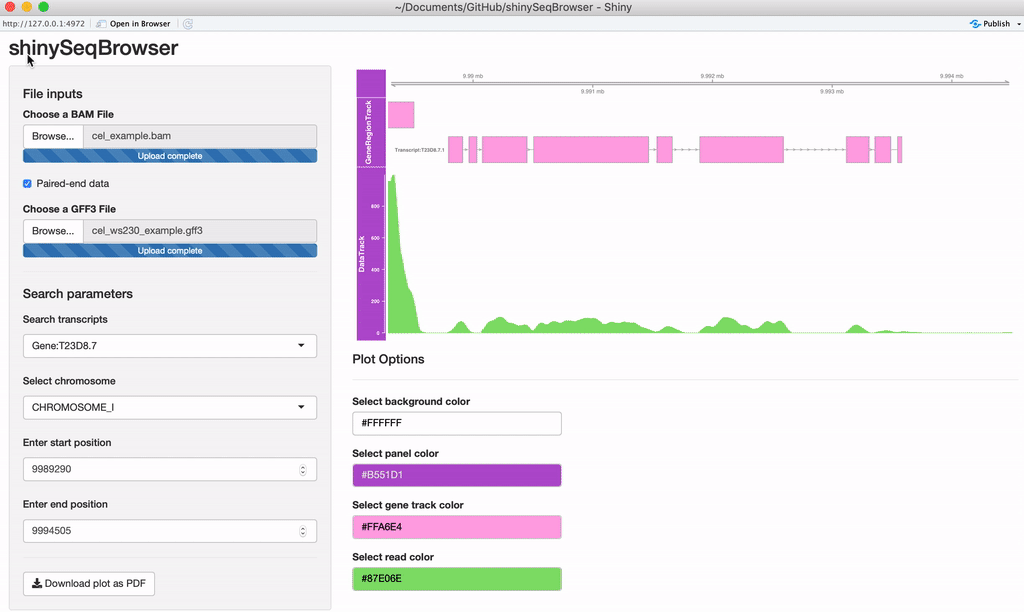
Upload your data: Once the app is up and running, you'll need to upload an aligned, sorted BAM file and an annotation file (GFF3/GTF). If the data is paired-end, you'll want to check that box as well.
Search Parameters: Once the annotation file is uploaded, the chromosomes and search options should appear. If you recieve an error here then something is wrong with your GFF file and GenomicFeatures package cannot parse it for some reason.
A plot should load after everything is uploaded. Every time a modification is made the plot will reload and a loading animation will appear to indicate it is updating (thanks to shinycssloader!). So if you do not see this animation, nor the plot, nor an error, something may be wrong. From here you can search a transcript by name or browse a chromosome by region.
Modify plot colors: You can modify the plot coloring using the color palette of your choice, colourpicker allows you to pick a color from a palette or enter the value so you can match the color schemes of your figures instead of having to modify this in post-processing.
Exporting: Once you're happy with it, you can save the plot as a vector-editable PDF!
I do not have contributing guidelines for this repository yet, but if you would like to contribute to the code feel free to fork the repository and create a new branch for your feature, bug fix, etc then create a pull request. Collaborative draft pull requests also welome. Create an issue if you have run into a bug or have a suggestion. Thanks!
To give you an idea of what I am working on next or where you might be able to contribute, here are the ideas I have that I would like to add as time goes on. This is very much a work in progress!
- Testing different files, finding bugs, and fixing them :)
- Upload genome & view sequence base information
- Split stranded reads
- Convert start/stop input to a slider range
- Compare >1 BAM file
- Add multiple plot styles available through
Gviz - Collapse and uncollapse transcripts/isoforms
- Load a UCSC genome
- More plot modification options: text, scale
- Kristen Brown (biokcb) - Colorado State University
See also the list of contributors for more on who has participated.
Due to R shiny being licensed GPLv3, I believe this app must also be licensed GPLv3, but it is unclear to me.. clarifications welcome. :) See the LICENSE.
As this was a new set of tools and learning experience for me, there was quite a lot of reading and searching involved and it would have taken significantly more time and effort to produce without these resources. To create this I mainly followed the Gviz user guide, the Shiny tutorials, function descriptions and community page, good ol' stackoverflow, and some useful articles from Dave Tang's blog. GIFs captured with Kap.