Quick Links
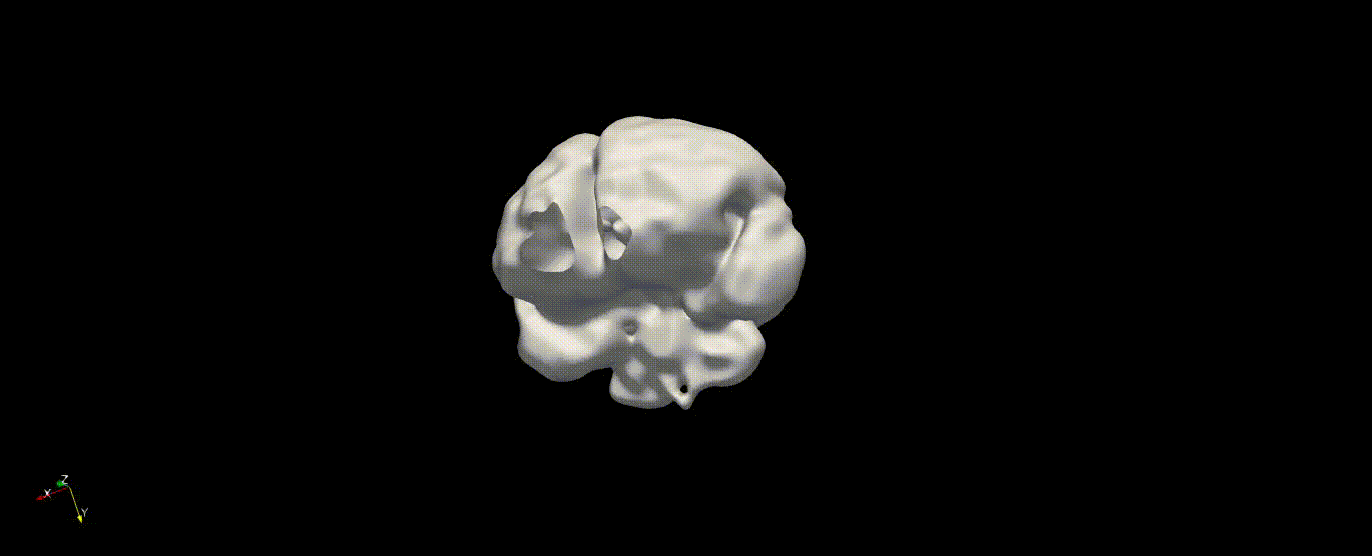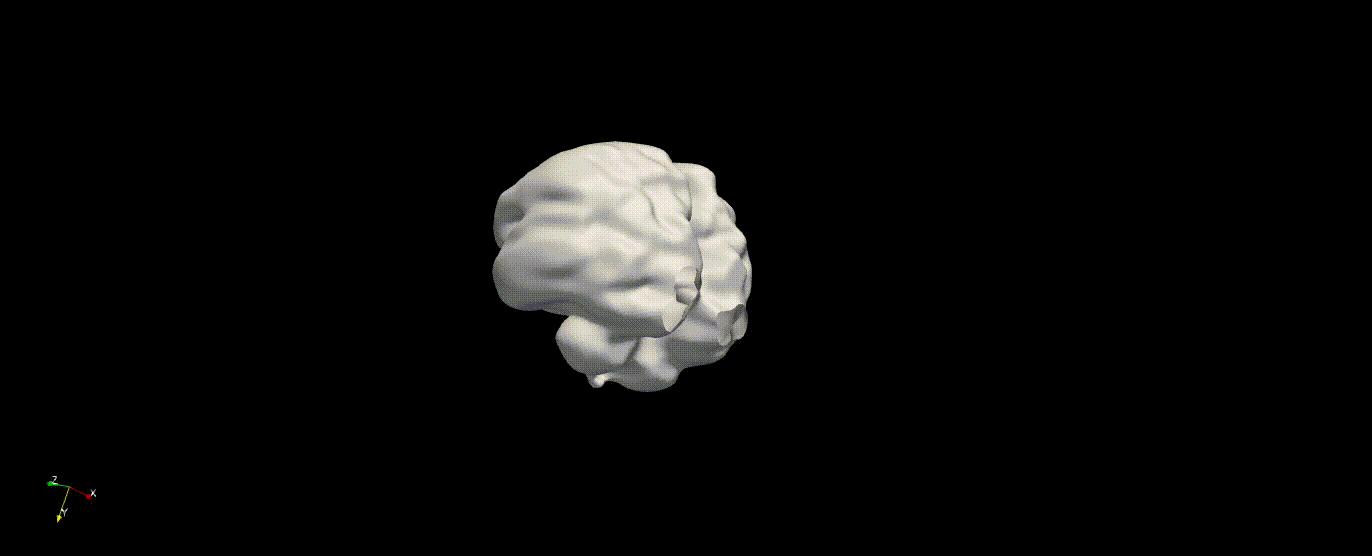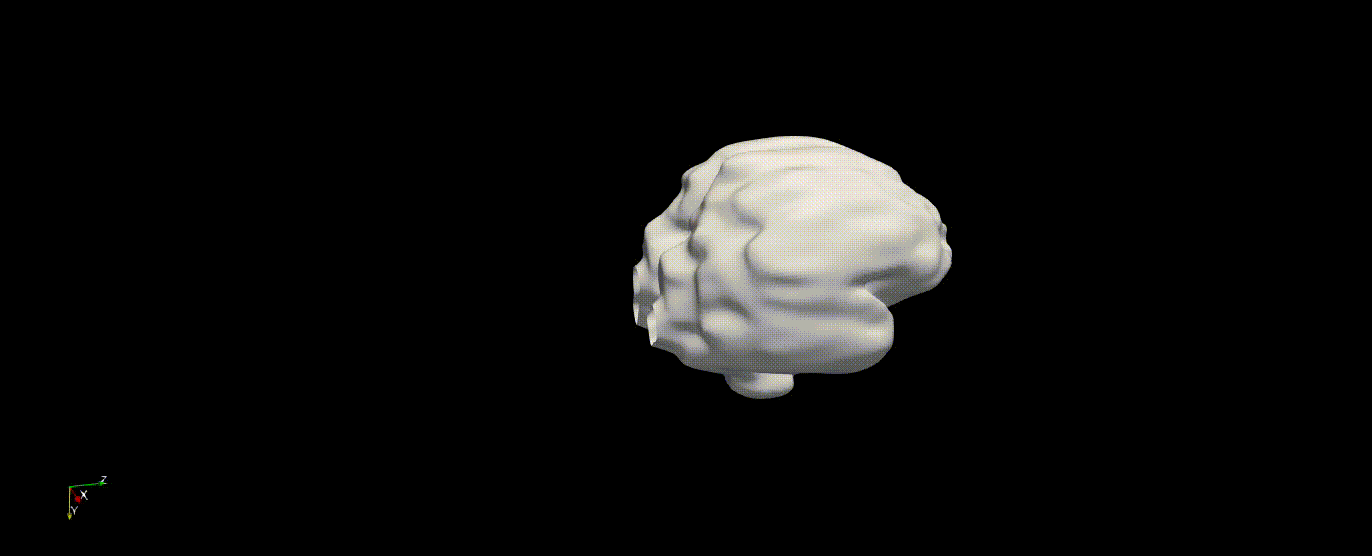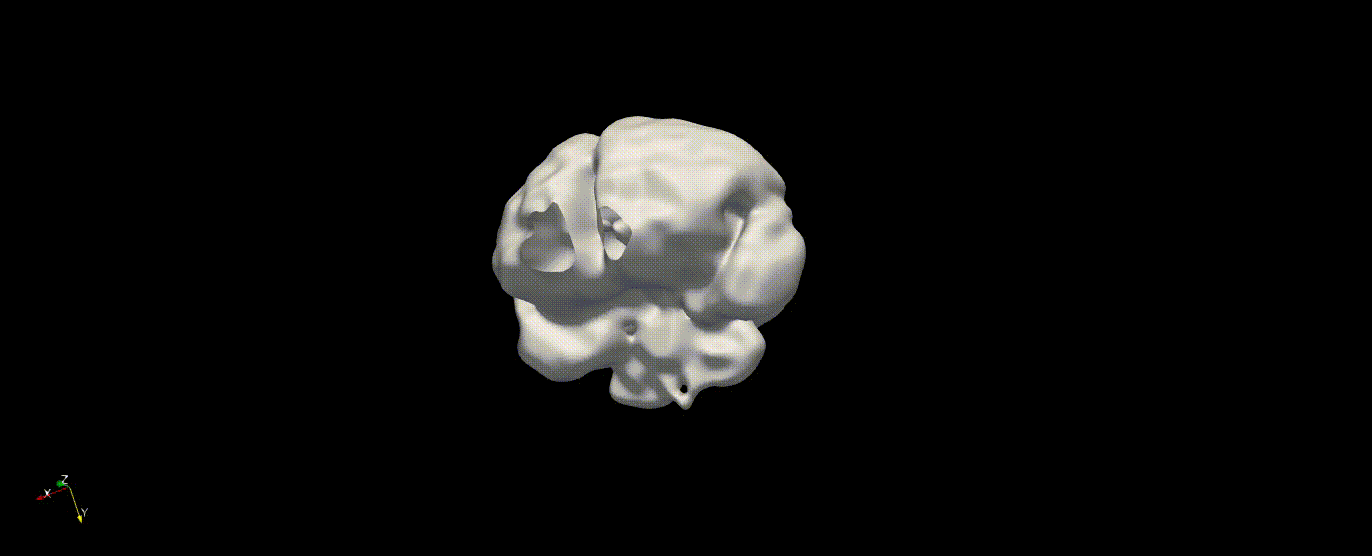
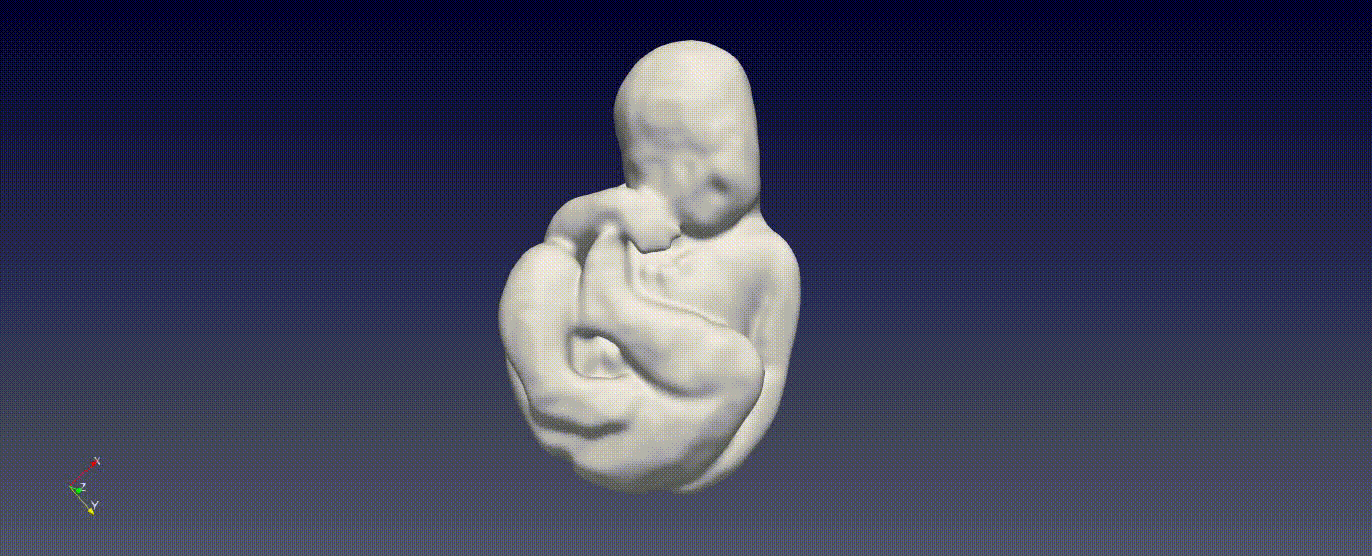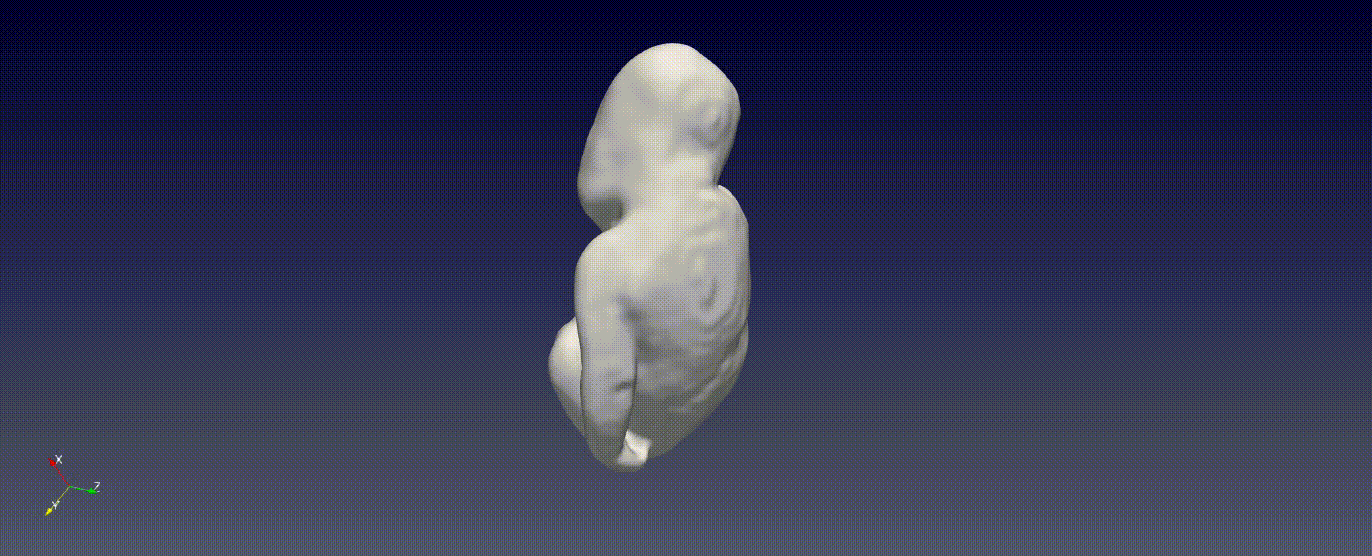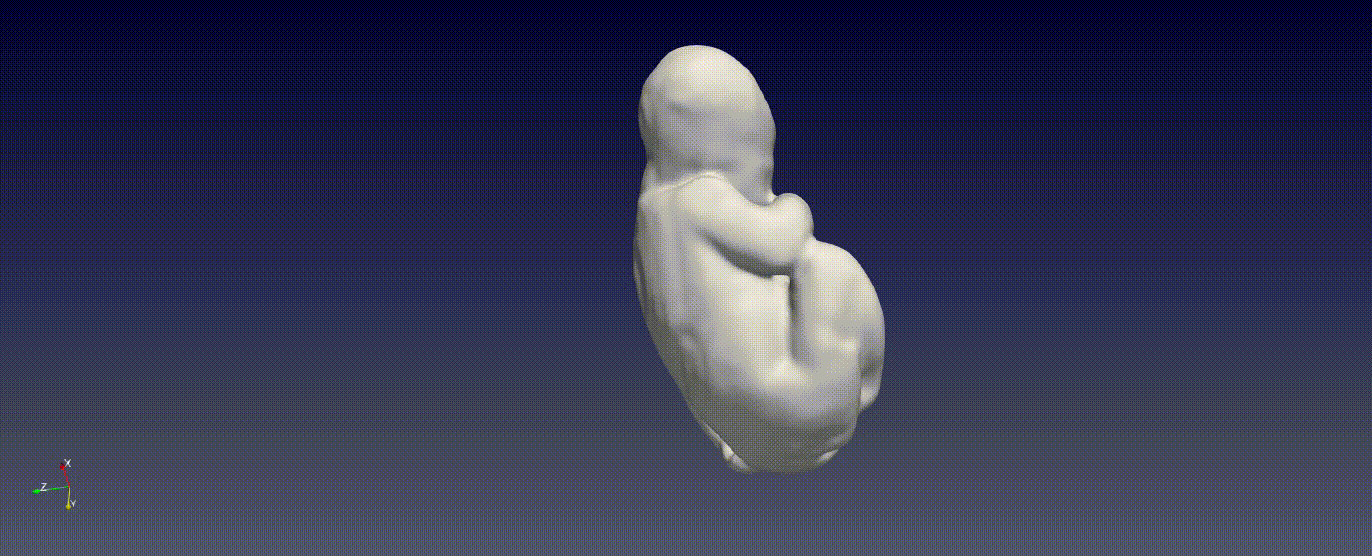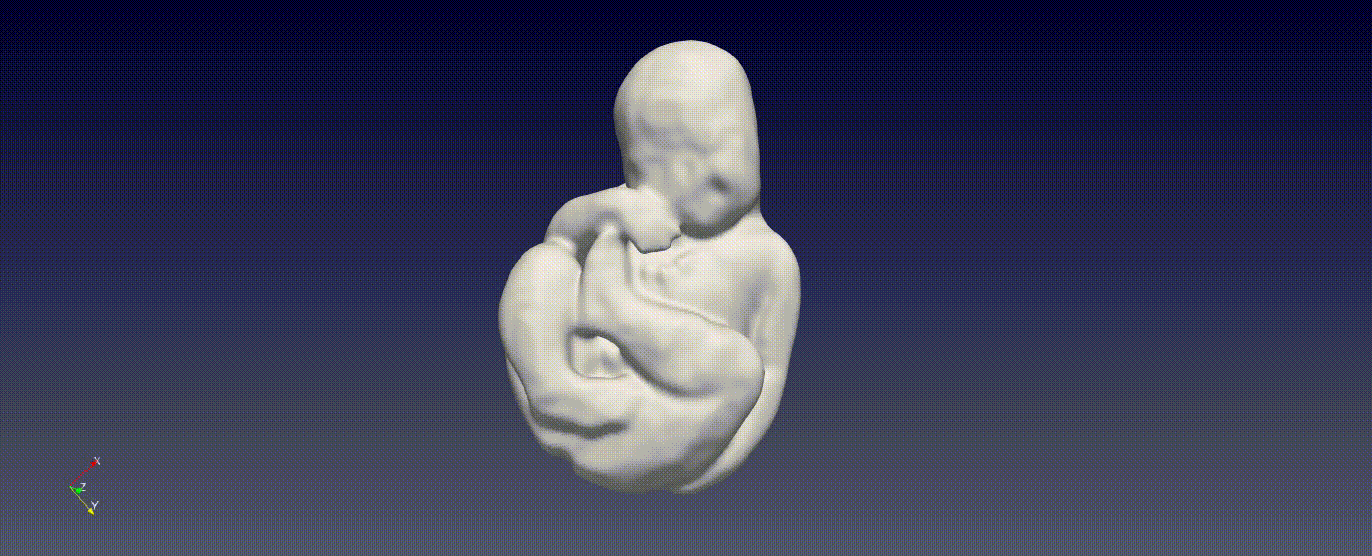
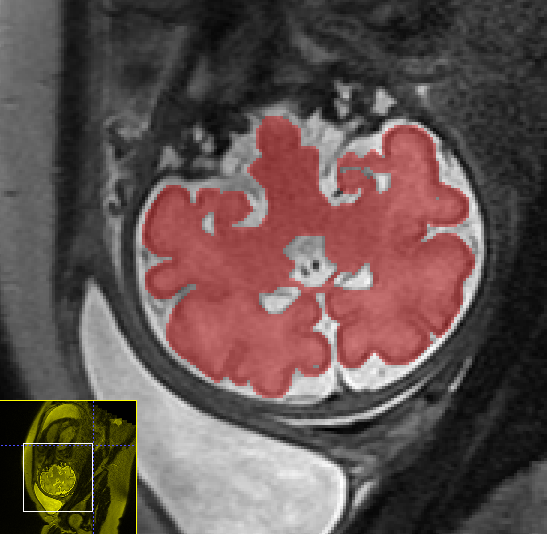
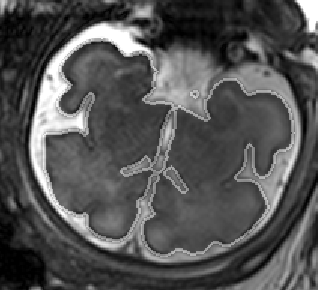
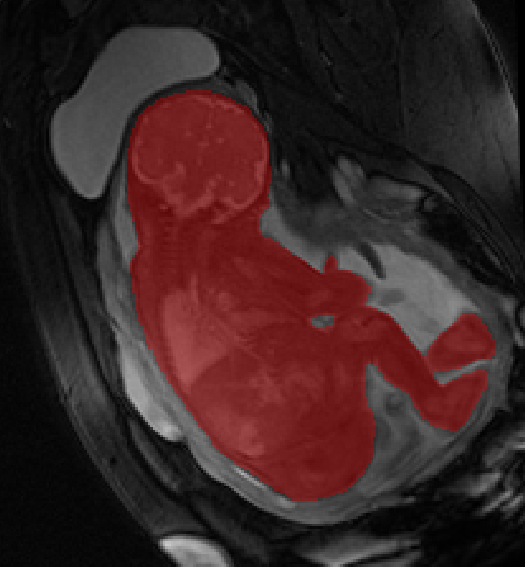
Fetal Total-Body and Brain 3D Segmentation in MRI scans with limited datasets.
- 3D and 2D segmentation.
- Implemented Advanced Augmentation techniques.
- Achieving results below Radiologist Inter & Intra Observer variability.
- Option to Utilize previous-slice segmentation to improve performance on hard samples.
Training
-
Install Tensorflow & Keras
-
Install dependencies: pip install -r requirements.txt
-
To run training: Config currently in dict inside brats/train_fetal.py
$ python -m brats.train_fetal --config_dir <config_dir>
Where <config_dir> is the folder containing the training configurations (or will be creatad to)
Write prediction images from the validation data
In the training above, part of the data was held out for validation purposes. To write the predicted label maps to file:
$ python -m brats.predict --config_dir <config_dir>
The predictions will be written in the <config_dir>/prediction folder along with the input data and ground truth labels for comparison.