Deep Learning-Based Object Detection and Bacteria Morphological Feature Extraction for Antimicrobial Resistance Applications
See our paper on IEEEXplore.
Antibiotics are the primary drug for treating various kinds of infections occurring from bacteria and microbes. They work mainly by blocking the vital pathway of those organisms and stopping them from multiplying. Previous research shows that we can predict the antibiotics used on bacteria by visualizing their morphology. Here, we present object detection for detecting bacteria and identifying the antibiotics used on them with their mophological features for example DNA intensity, contour area, and min areaRect.
There are 8 common classes we are interested including E.Coli bacteria treated with Ampicillin, Ciprofloxacin, Rifampicin, Tetracycline, Mecillinam, Kanamycin, Colistin, and Untreated.
The current results of single models and ensemble models are as follows.
| Model | Backbone | Head | Neck | mAP | mIOU | AP(50) | AP(75) | AP (medium) | AP (large) | Config | Checkpoint |
|---|---|---|---|---|---|---|---|---|---|---|---|
| YOLOv2 | Darknet-19 | 0.053 | 0.192 | 0.015 | 0.048 | 0.102 | 0.140 | config | ckpt | ||
| Faster R-CNN | ResNet-50 | 0.041 | 0.097 | 0.031 | 0.005 | 0.045 | 0.325 | config | ckpt | ||
| Cascade R-CNN | Res2Net-101 + DCNv2 | SABL | PAFPN + DyHead | 0.652 | 0.800 | 0.808 | 0.762 | 0.677 | 0.692 | config | ckpt |
| YOLOX-M | CSPDarknet | - | PAFPN | 0.621 | 0.755 | 0.902 | 0.835 | 0.711 | 0.796 | config | ckpt |
| Cascade R-CNN | Res2Net-50 + DCNvV2 | SABL | PAFPN + DyHead | 0.680 | 0.802 | 0.820 | 0.779 | 0.704 | 0.628 | config | ckpt |
| Ensemble Model | 0.753 | 0.699 | 0.863 | 0.796 | 0.717 | 0.675 |
We apply the following approach to extract bacteria features:
- Feature Pyramid Network (FPN) for automatic color manipulation
- Deep MAC for instance segmentation from the bounding box given by object detection models
- Open-CV for feature extraction. We extract 19 morphological features using OpenCV to obtain the perimeter, area, length, and width of the cell membrane, DNA, and color intensity (minimum, maximum, mean, median, and standard deviation of the green and blue color channels)
We compare downstream antibiotic classification with SVM using features extracted from our model and CellProfiler.
| Bacteria detection model | CellProfiler | |
|---|---|---|
| Mean F1-score | 0.76 | 0.796 |
This difference is considered acceptable because the number of bacteria that the CellProfiler can detect is significantly lower than the model’s but higher in terms of quality since the CellProfiler can only detect complete bacteria cells.
The current dataset contains 900 images: Ampicillin (100), Ciprofloxacin (100), Rifampicin (100), Tetracycline (100), Mecillinam(100), Kanamycin(100), Colistin(100), and Untreated (200). We annotated the RGB version in PNG format of the bacteria images which is easier to visualize using labelme. Images are in TIFF format that we use for actual model training and testing.
| High intensity image | Low intensity image |
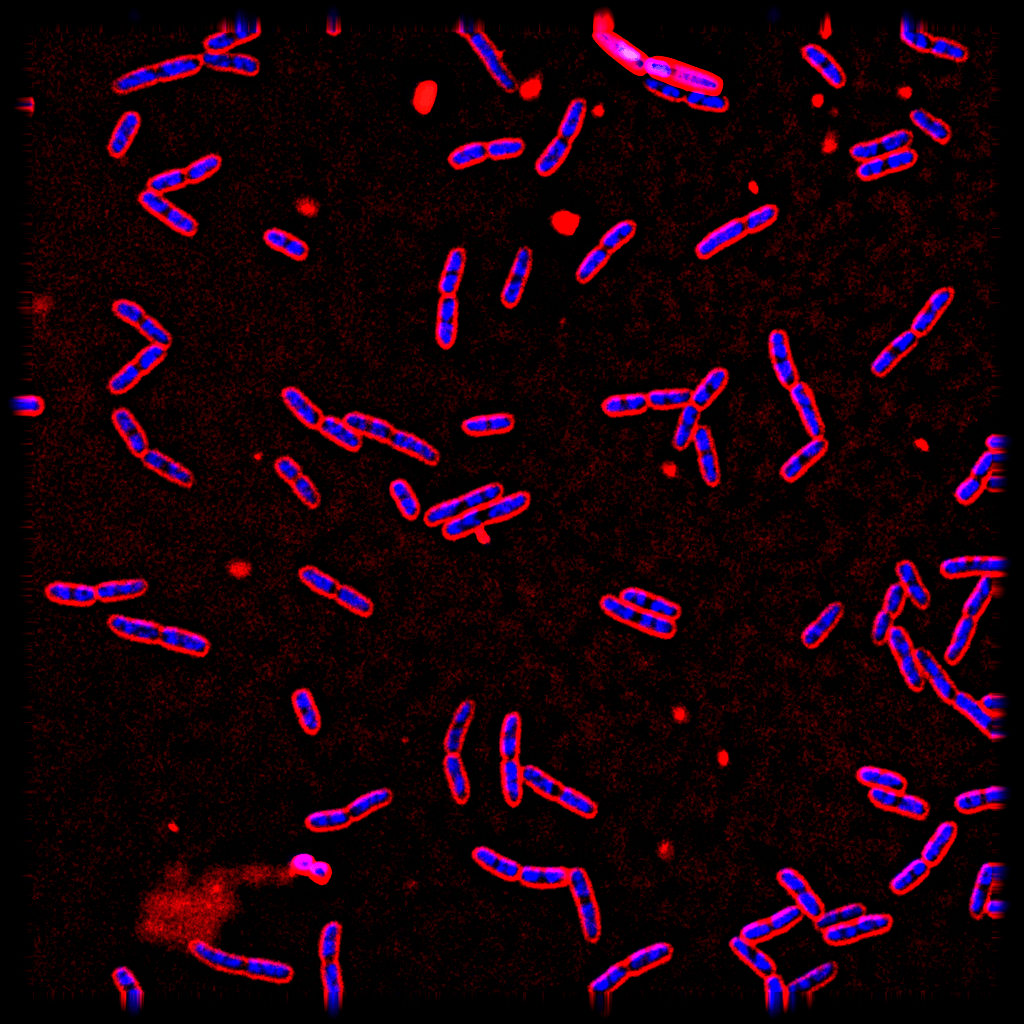
|

|
- Download the virtual environment from here
- Move the virtual environment to the bacteria-detection folder
- extract the virtual environment
cd bacteria-detection
conda activate ./path/to/virtual_environmentWe release the pre-trained model weight for reproducibility purposes. You can download the weights of all models by DVC
dvc pullcd webapp/backend
uvicorn app:app --reloadWe use ReactJS as our frontend. To run an application, install NodeJS here and run the following
cd webapp/frontend/bacteria-app
npm startcd webapp/backend/inference
python inference_ensemble.py --imgs_folder <Path/to/Image/Folder>The outputs will be in the output folder and include
- CSV file of all detection results
- CSV file of all extracted features
YOLOv2 model is trained using darknet
Other object detection models are trained using mmdetection
If you use our project or the our paper, please cite as
@INPROCEEDINGS{10322010,
author={Chotayapa, Korrawiz and Leethamchayo, Thanyatorn and Chinnawong, Piraya and Samernate, Thanadon and Nonejuie, Poochit and Achakulvisut, Titipat},
booktitle={2023 15th Biomedical Engineering International Conference (BMEiCON)},
title={Deep Learning-Based Object Detection And Bacteria Morphological Feature Extraction For Antibiotic Mode Of Action Study},
year={2023},
volume={},
number={},
pages={1-5},
doi={10.1109/BMEiCON60347.2023.10322010}}
We would like to thank Poochit Nonejuie Ph.D. and Mr. Thanadon Samernate from the Institute of Molecular Biosciences that inspired us and prepared the dataset for this study.



