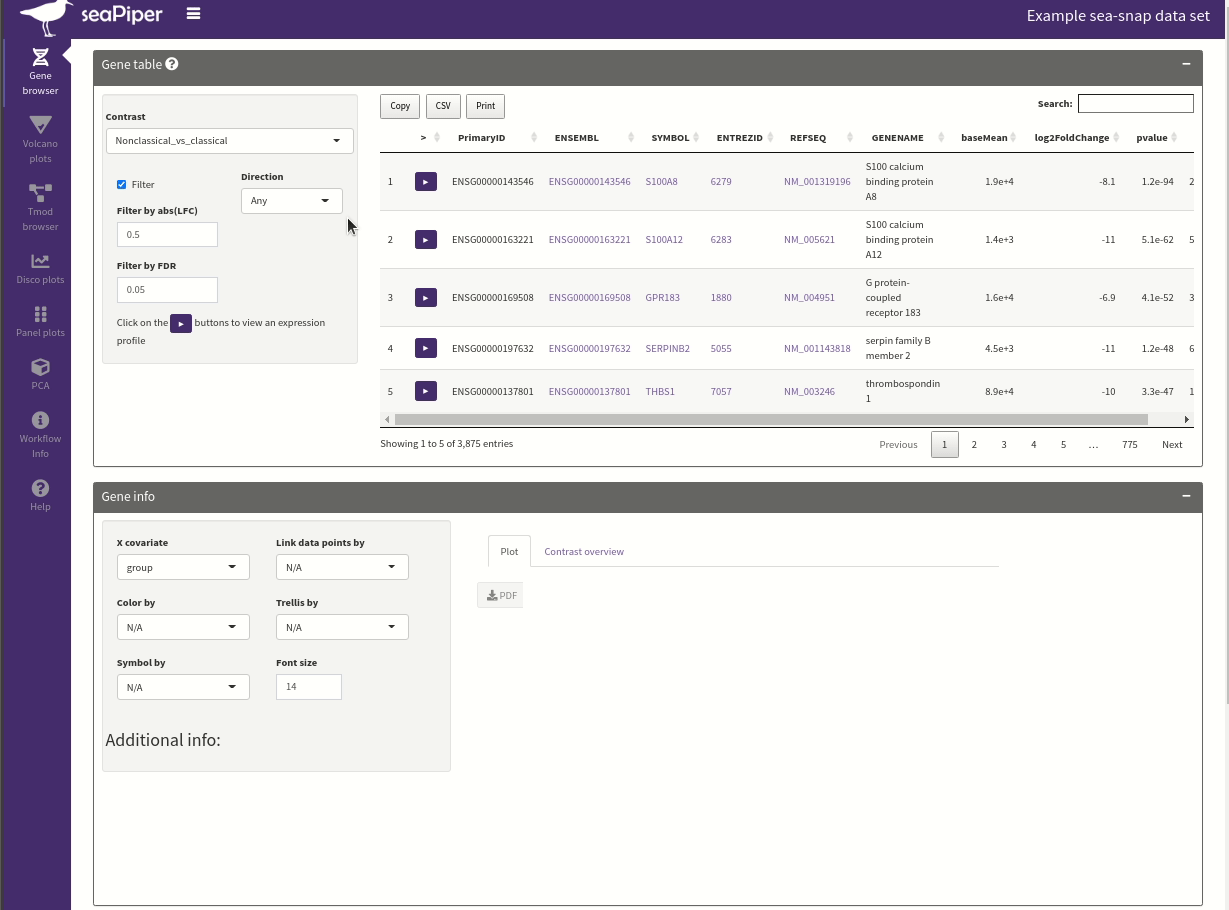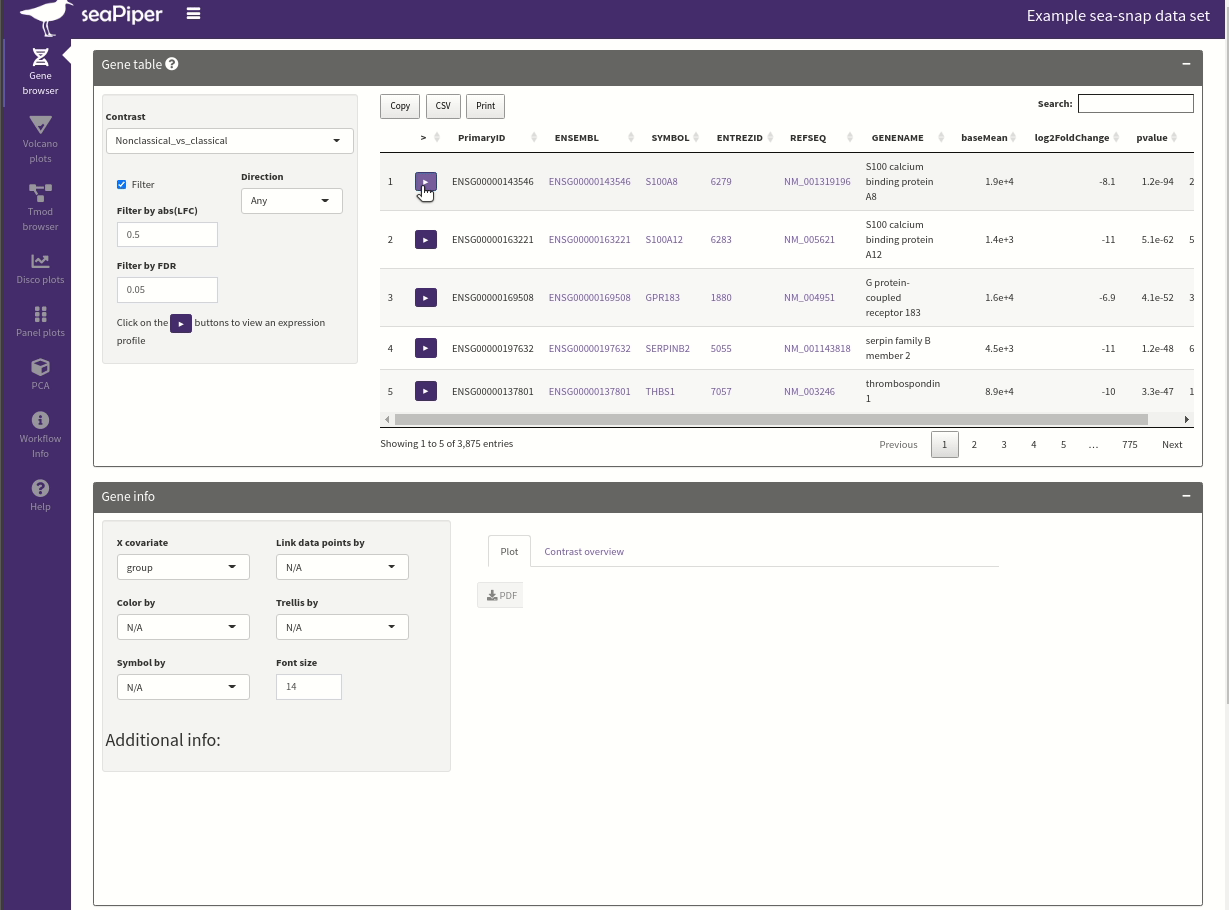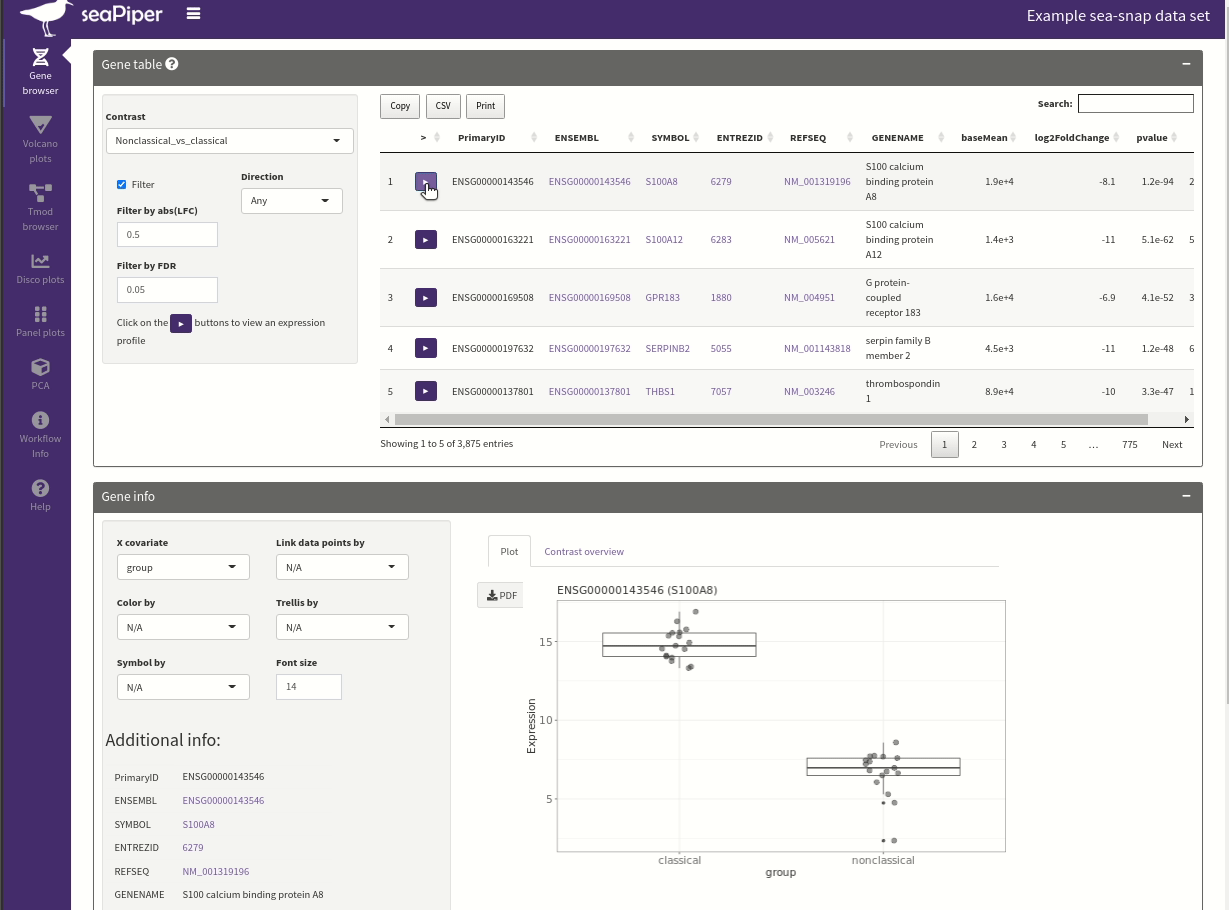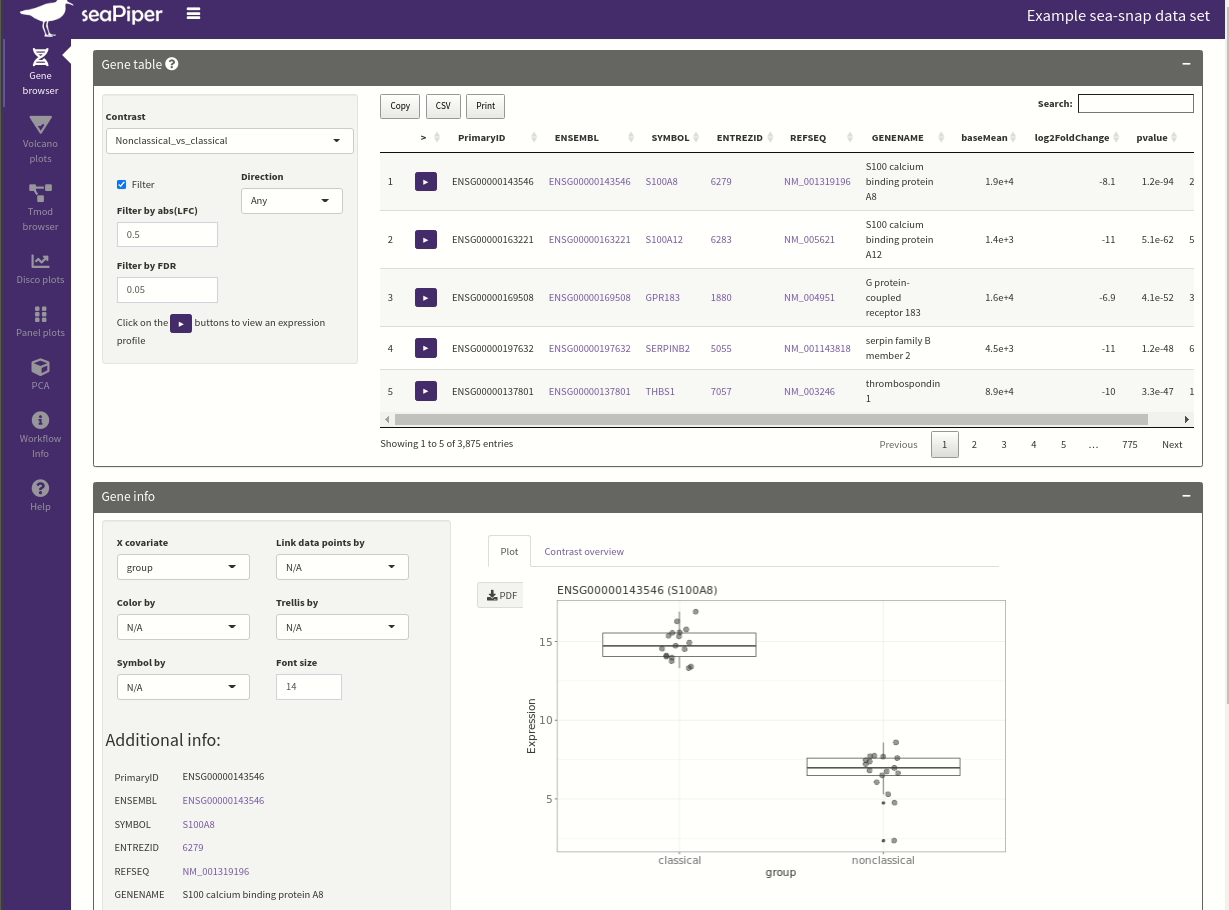
seaPiper 
Workflow output explorer for the sea-snap pipeline
Installation
You can install seaPiper from GitHub with:
# install.packages("devtools")
devtools::install_github("bihealth/seaPiper")Example
This is a basic example which shows you how to solve a common problem:
library(seaPiper)
## basic example code