- Benjamin Fair (@bfairkun)
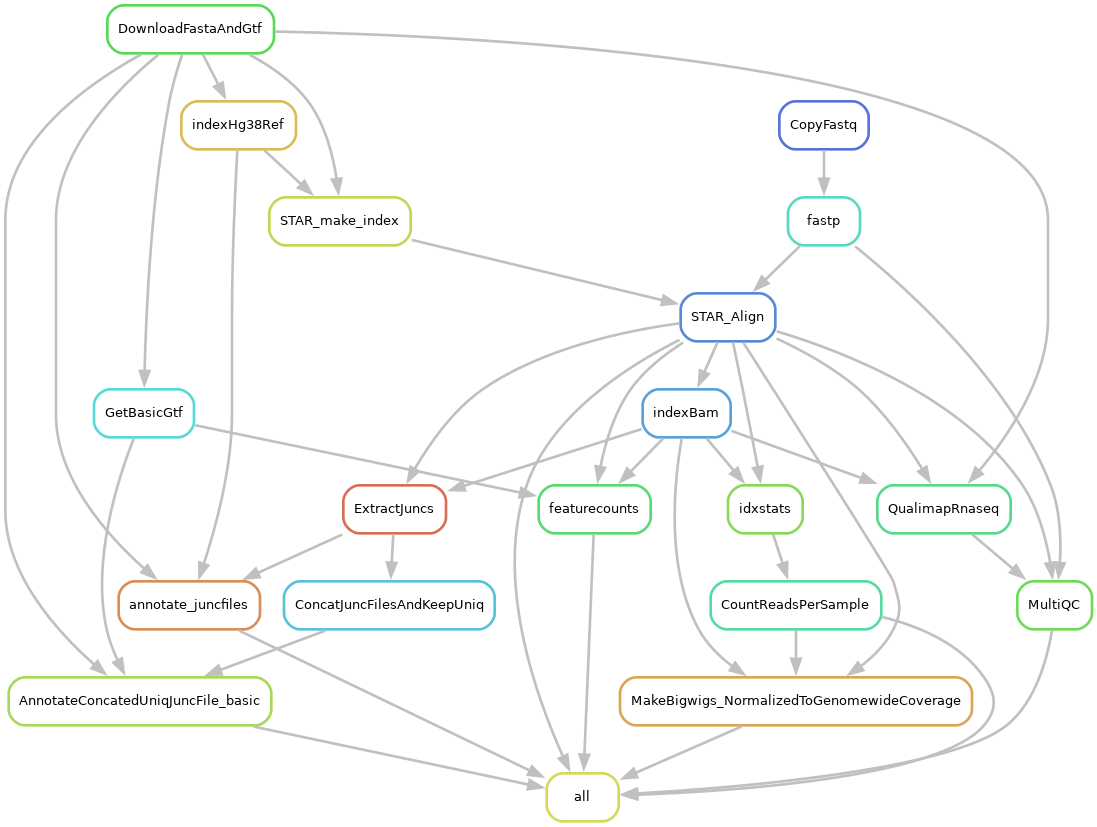
This workflow contains rules to download genome files, index genome and align reads with STAR, perform some basic QC, count splice junction reads (regtools) and gene reads (feature counts). Can handle different samples from different species, as defined in config/STAR_Genome_List.tsv and config/samples.tsv. Because this is often just the start of a RNA-seq analysis, this workflow might be best used as a module in a Snakemake workflow that further extends this work.
If you simply want to use this workflow, clone the latest release.
git clone git@github.com:bfairkun/rna-seq_simplequantification.git
If you intend to modify and further develop this workflow, fork this repository. Please consider providing any generally applicable modifications via a pull request.
Install snakemake and the workflow's other dependencies via conda/mamba. If conda/mamba isn't already installed, I recommend installing miniconda and then install mamba in your base environment. Then...
# move to the snakemake's working directory
cd rna-seq_simplequantification/code
# Create environment for the snakemake
mamba env create -f envs/rna-seq_simplequantification.yaml
# And activate the enviroment
conda activate rna-seq_simplequantification
Configure the workflow according to your needs via editing the file config.yaml. Use/modify the config yaml files in the snakemake_profiles/slurm/ profile to run on UChicago RCC Midway with slurm scheduler.
Test your configuration by performing a dry-run via
snakemake -n
Execute the workflow locally via
snakemake --cores $N
using $N cores or run it in a cluster environment via the included slurm snakemake profile.
snakemake --profile snakemake_profiles/slurm
See the Snakemake documentation for further details.