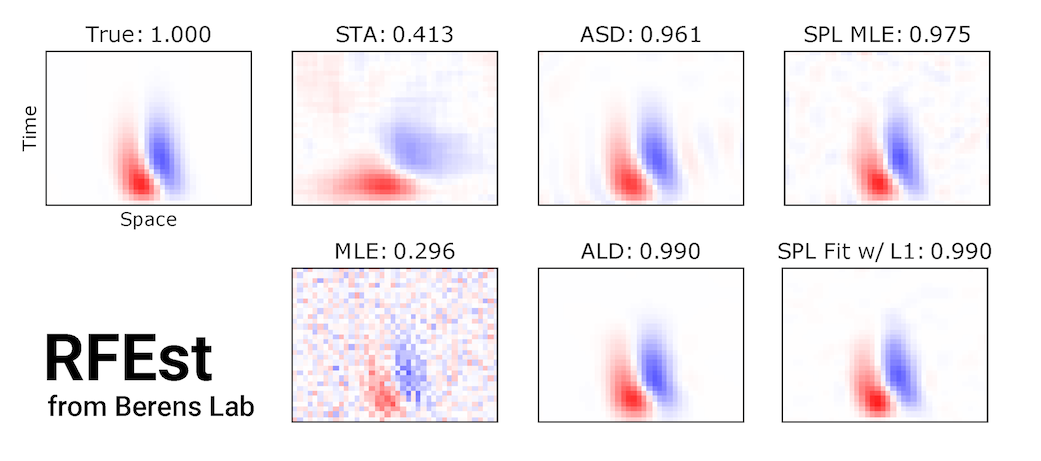
RFEst v2 is a Python3 toolbox for neural receptive field estimation, featuring methods such as spline-based GLMs, Empirical Bayes with various Gaussian priors, and a few matrix factorization methods.
Spline-based GLMs [1]
The new GLM module unified both vanilla and spline GLMs.
from rfest import GLM
lnp = GLM(distr='poisson', output_nonlinearity='softplus')
# add training data
lnp.add_design_matrix(X_train, dims=[25, ], df=[8, ], smooth='cr', name='stimulus') # use spline for stimulus filter
lnp.add_design_matrix(y_train, dims=[20, ], df=[8, ], smooth='cr', shift=1,
name='history') # use spline for history filter
# add validation data
lnp.add_design_matrix(X_dev, name='stimulus') # basis will automatically apply to dev set
lnp.add_design_matrix(y_dev, name='history')
# intialize model parameters
lnp.initialize(num_subunits=1, dt=dt, method='random', random_seed=2046)
# fit model
lnp.fit(y={'train': y_train, 'dev': y_dev},
num_iters=1000, verbose=100, step_size=0.1, beta=0.01)Evidence Optimization
- Ridge Regression
- Automatic Relevance Determination (ARD) [2]
- Automatic Smoothness Determination (ASD) [3]
- Automatic Locality Determination (ALD) [4]
from rfest import ASD
asd = ASD(X, y, dims=[5, 20, 15]) # nT, nX, nY
p0 = [1., 1., 2., 2., 2.] # sig, rho, 𝛿t, 𝛿y, 𝛿x
asd.fit(p0=p0, num_iters=300)Matrix Factorization
A few matrix factorization methods have been implemented as a submodule (MF).
from rfest.MF import KMeans, semiNMFFor more information, see here.
RFEst uses JAX for automatic differentiation and JIT compilation to GPU/CPU, so you need to install JAX first.
To install CPU-only version for Linux and macOS, simply clone this repo into a local directory and install
via pip:
git clone https://github.com/berenslab/RFEst
pip install -e RFEstTo enable GPU support on Linux, you need to consult the JAX install guide. For reference purpose, I copied the relevant steps here, but please always check the JAX README page for up-to-date information.
# install jaxlib
PYTHON_VERSION=cp37 # alternatives: cp36, cp37, cp38
CUDA_VERSION=cuda100 # alternatives: cuda100, cuda101, cuda102, cuda110
PLATFORM=manylinux2010_x86_64 # alternatives: manylinux2010_x86_64
BASE_URL='https://storage.googleapis.com/jax-releases'
pip install --upgrade $BASE_URL/$CUDA_VERSION/jaxlib-0.1.50-$PYTHON_VERSION-none-$PLATFORM.whl
pip install --upgrade jax # install jaxJAX has no native Windows support yet, but can be installed on CPU via the Windows Subsystem for Linux (Windows 10
only, and make sure that the pip version is the latest pip install --upgrade pip).
numpy
scipy
sklearn
matplotlib
jax
jaxlib
Tutorial notebooks can be found here: https://github.com/huangziwei/notebooks_RFEst
[1] Huang, Z., Ran, Y., Oesterle, J., Euler, T., & Berens, P. (2021). Estimating smooth and sparse neural receptive fields with a flexible spline basis. Neurons, Behavior, Data Analysis, and Theory, 5(3), 1–30. https://doi.org/10.51628/001c.27578
[2] MacKay, D. J. (1994). Bayesian nonlinear modeling for the prediction competition. ASHRAE transactions, 100(2), 1053-1062.
[3] Sahani, M., & Linden, J. F. (2003). Evidence optimization techniques for estimating stimulus-response functions. In Advances in neural information processing systems (pp. 317-324).
[4] Park, M., & Pillow, J. W. (2011). Receptive field inference with localized priors. PLoS computational biology, 7(10) , e1002219.