Pippin - a pipeline designed to streamline and remove as much hassle as we can when running end-to-end supernova cosmology analyses.
The (work in progress) docs can be found here.
- Using Pippin
- Installing Pippin
- Contributing to Pippin
- Examples
- FAQ
- Tasks
- Adding a new Task
- Adding a new classifier
If you're using a pre-installed version of Pippin - like the one on Midway, ignore this.
If you're not, installing Pippin is simple.
- Checkout Pippin
- Ensure you have the dependencies install (
pip install -r requirements.txt) and that your python version is 3.7+. - Celebrate
There is no need to attempt to install Pippin like a package (no python setup.py install), just run from the clone.
Now, Pippin also interfaces with other tasks: SNANA and machine learning classifiers mostly. I'd highly recommend running on a high performance computer with SNANA already installed, but if you want to take a crack at installing it, you can find the docoumentation here.
I won't cover installing SNANA here, hopefully you already have it. But to install the classifiers, we'll take SuperNNova as an example. To install that, find a good place for it and:
- Checkout
https://github.com/SuperNNova/SuperNNova - Create a GPU conda env for it:
conda create --name snn_gpu --file env/conda_env_gpu_linux64.txt - Activate environment and install natsort:
conda activate snn_gpuandconda install --yes natsort
Then, in the Pippin global configuration file cfg.yml in the top level directory, ensure that the SNN path in Pippin is
pointing to where you just cloned SNN into. You will need to install the other external software packages
if you want to use them, and you do not need to install any package you do not explicitly request in a config file.
Using Pippin is very simple. In the top level directory, there is a pippin.sh. If you're on midway and use SNANA, this
script will be on your path already. To use Pippin, all you need is a config file ready to go. I've got a bunch of mine and
some general ones in the configs directory, but you can put yours wherever you want. I recommend adding your initials to the
front of the file to make it obvious in the shared output directory which folders as yours.
If you have example.yml as your config file and want pippin to run it, easy:
pippin.sh example.yml
The file name that you pass in should contain a run configuration. Note that this is different to the global software
configuration file cfg.yml, and remember to ensure that your cfg.yml file is set up properly and that you know
where you want your output to be
installed. By default, I assume that the $PIPPIN_OUTPUT environment variable is set as the output location,
so please either set said variable or change the associated line in the cfg.yml. For the morbidly curious, here
is a very small demo video of using Pippin in the Midway environment.
Each configuration file is represented by a yaml dictionary linking each stage (see stage declaration section below) to a dictionary of tasks, the key being the unique name for the task and the value being its specific task configuration.
For example, to define a configuration with two simulations and one light curve fitting task (resulting in 2 output simulations and 2 output light curve tasks - one for each simulation), a user would define:
SIM:
SIM_NAME_1:
SIM_CONFIG: HERE
SIM_NAME_2:
SIM_CONFIG: HERE
LCFIT:
LCFIT_NAME_1:
LCFIT_CONFIG: HEREHow to configure each task is also detail below on a task-by-task basis, or you can see examples in the examples
directory for each task.
Happens all the time, don't even worry about it. Just start Pippin again and run the file again. Pippin will detect
any changes in your configuration by hashing all the input files to a specific task. So this means, even if you're
config file itself doesn't change, changes to an input file it references (for example, the default DES simulation
input file) would result in Pippin rerunning that task. If it cannot detect anything has changed, and if the task
finished successfully the last time it was run, the task is not re-executed. You can force re-execution of tasks using the -r flag.
On top of this, Pippin has a few command line arguments, which you can detail with pippin.sh -h, but I'll also detail here:
-h Show the help menu
-v, --verbose Verbose. Shows debug output. I normally have this option enabled.
-r, --refresh Refresh/redo - Rerun tasks that completed in a previous run even if the inputs haven't changed.
-c, --check Check that the input config is valid but don't actually run any tasks.
-s, --start Start at this task and refresh everything after it. Number of string accepted
-f, --finish Finish at this stage. For example -f 3 or -f CLASSIFY to run up to and including classification.
-p, --permission Fix permissions and groups on all output, don't rerun
-i, --ignore Do NOT regenerate/run tasks up to and including this stage.
-S, --syntax If no task is given, prints out the possible tasks. If a task name or number is given, prints the docs on that task. For instance 'pippin.sh -S 0' and 'pippin.sh -S DATAPREP' will print the documentation for the DATAPREP task.For an example, to have a verbose output configuration run and only do data preparation and simulation, you would run
pippin.sh -vf 1 configfile.yml
You may have noticed above that each stage has a numeric idea for convenience and lexigraphical sorting.
The current stages are:
0, DATAPREPData preparation1, SIM: Simulation2, LCFIT: Light curve fitting3, CLASSIFY: Classification (training and testing)4, AGG: Aggregation (comparing classifiers)5, MERGE: Merging (combining classifier and FITRES output)6, BIASCOR: Bias corrections using BBC7, CREATE_COV: Create input files needed for CosmoMC8, COSMOFIT: Run CosmoMC and fit cosmology9, ANALYSE: Create final output and plots. Includes output from CosmoMC, BBC and Light curve fitting.
On midway, sourcing the SNANA setup will add environment variables and Pippin to your path.
Pippin itself can be found at $PIPPIN, output at $PIPPIN_OUTPUT (which goes to a scratch directory), and pippin.sh will automatically work from
any location.
Note that you only have 100 GB on scratch. If you fill that up and need to nuke some files, look both in $SCRATCH_SIMDIR to remove SNANA
photometry and $PIPPIN_OUTPUT to remove Pippin's output. I'd recommend adding this to your ~/.bashrc file to scan through directories you own and
calculate directory size so you know what's taking the most space. After adding this and sourcing it, just put dirusage into the terminal
in both of those locations and see what's eating your quota.
function dirusage {
for file in $(ls -l | grep $USER | awk '{print $NF}')
do
du -sh "$file"
done
}Contributing to Pippin or raising issues is easy. Here are some ways you can do it, in order of preference:
- Submit an issue on Github, and then submit a pull request to fix that issue.
- Submit an issue on Github, and then wait until I have time to look at it. Hopefully thats quickly, but no guarantees.
- Email me with a feature request
If you do want to contribute code, fantastic. Please note that all code in Pippin is subject to the Black formatter. I would recommend installing this yourself because it's a great tool.
If you want detailed examples of what you can do with Pippin tasks, have a look in the examples directory, pick the task you want to know more about, and have a look over all the options.
Here is a very simple configuration file which runs a simulation, does light curve fitting, and then classifies it using the debug FITPROB classifier.
SIM:
DESSIM:
IA_G10_DES3YR:
BASE: surveys/des/sim_ia/sn_ia_salt2_g10_des3yr.input
LCFIT:
BASEDES:
BASE: surveys/des/lcfit_nml/des_5yr.nml
CLASSIFICATION:
FITPROBTEST:
CLASSIFIER: FitProbClassifier
MODE: predictYou can see that unless you specify a MASK on each subsequent task, Pippin will generally try and run everything on everything. So if you have two
simulations defined, you don't need two light curve fitting tasks, Pippin will make one light curve fit task for each simulation, and then two classification tasks,
one for each light curve fit task.
If you are finding that your config files contain lots of duplicated sections (for example, many simulations configured
almost the same way but with one differnece), consider using YAML anchors. See this blog post
for more detail. You can define your anchors in the main config section, or add a new section (like SIM, LCFIT, CLASSIFICATION). So long as it doesn't
match a Pippin keyword for each stage, you'll be fine. I recommend ANCHORS: at the top of the file, all of those will work.
Pippin is crashing on some task and the error message isn't useful
Feel free to send me the log and stack, and I'll see what I can do turn the exception into something more human-readable.
I want Pippin to run after I log out
Rather than redirecting Pippin output to a file or running it in the background, I highly recommend you run
Pippin in a screen session.
For example, if you are doing machine-learning testing, you may create a new screen session called ml
by running screen -S ml. It will then launch a new instance of bash for you to play around in. conda will not work out of the box. To make
it work again, run conda deactivate and then conda activate, and you can check this works by running which python and
verifying its pointing to the miniconda install. You can then run Pippin as per normal: pippin.sh -v your_job.yml and get the coloured output.
To leave the screen session, but still keep Pippin running even after you log out, press Ctrl-A, Ctrl-D. As in one, and then the other, not Ctrl-A-D.
This will detach from your screen session but keep it running. Just going Ctrl_D will disconnect and shut it down. To get back into your screen session,
simply run screen -r ml to reattach. You can see your screen
sessions using screen -ls.
You may notice if you log in and out of midway that your screen sessions might not show up. This is because midway has multiple head nodes, and
your screen session exists only on one of them. This is why when I ssh to midway I specify a specific login node instead
of being assigned one. To make it simpler, I'd recommend setting up
an alias like so to either login1 or login2:
alias sshmidway="ssh username@midway2-login1.rcc.uchicago.edu"I want to modify a ton of files but don't want huge yml files, please help
You can modify input files and put them in a directory you own, and then tell Pippin to look there (in addition to the default location) when its constructing your tasks. To do this, see this example here, or use this code snippet at the top of your YAML file (not that it matters if it's at the top):
GLOBAL:
DATA_DIRS:
- /some/new/directory/with/your/files/in/itI want to use a different cfg.yml file!
GLOBAL:
CFG_PATH: /your/path/hereStop rerunning my sims!
For big biascor sims it can be frustrating if you're trying to tweak biascor or later stages and sims kick off
because of some trivial change. So use the --ignore ro -i command to ignore any undone tasks or tasks with
hash disagreements in previous stages. To clarify, even tasks that do not have a hash, and have never been submitted, will
not be run if that stage is set to be ignored.
I don't want to run this massive jobs again! Let me use external results!
Good news, everyone! Not only is there a dedicated config file for globally useful tasks, but its easier than ever to slow them
into your existing jobs. For useful precomputed work, such as biascor sims and trained machine learning classifiers, check out $PIPPIN_OUTPUT/GLOBAL.
For an example on how to use these results, check out the reference 5YR analysis ref_des_5yr.yml. There are in essense two ways of
including external tasks. Both operate the same way, one is just a bit more explicit than the other. The explicit way is when adding
a task that is an exact replica of an external task, you can just add the EXTERNAL keyword. For example, in the reference 5YR analysis,
all the biascor sims are precomputed, so we can define them as external tasks like this:
SIM:
DESSIMBIAS5YRIA_C11:
EXTERNAL: $PIPPIN_OUTPUT/GLOBAL/1_SIM/DESSIMBIAS5YRIA_C11
DESSIMBIAS5YRIA_G10:
EXTERNAL: $PIPPIN_OUTPUT/GLOBAL/1_SIM/DESSIMBIAS5YRIA_G10
DESSIMBIAS5YRCC:
EXTERNAL: $PIPPIN_OUTPUT/GLOBAL/1_SIM/DESSIMBIAS5YRCCPoint to the external task output directory. In this case, we use the EXTERNAL keyword because each task we define has an explicit one-to-one match
with an external task.
But then say we don't want to recompute the light curve fits. After all, most of the time we're not changing that step anyway! Well, we set up the tasks in the same way as we normally would, but now there isn't a one-to-one mapping. See the config snippet below:
LCFIT:
D:
BASE: surveys/des/lcfit_nml/des_5yr.nml
MASK: DESSIM
EXTERNAL_DIRS:
- $PIPPIN_OUTPUT/GLOBAL/2_LCFIT/D_DESSIMBIAS5YRIA_C11
- $PIPPIN_OUTPUT/GLOBAL/2_LCFIT/D_DESSIMBIAS5YRIA_G10
- $PIPPIN_OUTPUT/GLOBAL/2_LCFIT/D_DESSIMBIAS5YRCCThat is, we have one LCFIT task, but because we have three sims going into it and matching the mask, we can't point a single external directory.
But what we can do is point to multiple! And then if the output directory matches (ie D_DESSIMBIAS5YRIA_C11 is the base name for both a computed
task output directory and an external task directory), it will instead use that.
Note that you still need to point to the right base file, because Pippin still wants those details. It won't be submitted anywhere though, just loaded in.
The flexibility of EXTERNAL_DIRS means you can mix both precomputed and non-precomputed tasks together. Take this classificaiton task:
CLASSIFICATION:
SNNTEST:
CLASSIFIER: SuperNNovaClassifier
MODE: predict
OPTS:
MODEL: $PIPPIN_OUTPUT/GLOBAL/3_CLAS/SNNTRAIN_DESTRAIN/model.pt
EXTERNAL_DIRS:
- $PIPPIN_OUTPUT/GLOBAL/3_CLAS/SNNTEST_DESSIMBIAS5YRIA_C11_SNNTRAIN_DESTRAIN
- $PIPPIN_OUTPUT/GLOBAL/3_CLAS/SNNTEST_DESSIMBIAS5YRIA_G10_SNNTRAIN_DESTRAIN
- $PIPPIN_OUTPUT/GLOBAL/3_CLAS/SNNTEST_DESSIMBIAS5YRCC_SNNTRAIN_DESTRAINIt will load in the precomputed classification results for the biascor sims, and then also run and generate classification results on any other
simulation tasks (such as running on the data) using the pretrained model model.pt.
Finally, the way this works under the hood is simple - it copies the directory over explicitly. And it will only copy once, so if you want the
"latest version" just ask the task to refresh (or delete the folder). Once it copies it, there is no normal hash checking,
it reads in the config.yml file created by the task in its initial run and powers onwards.
If you want to use EXTERNAL_DIRS with different output directory names, you will ned to inform pippin as to which EXTERNAL_DIR matches with which LCFIT task. To do so, you can make use of EXTERNAL_MAP, which allows you to specify a mask associated with each LCFIT task, selecting an EXTERNAL_DIR for that task. Note that tasks which do have an exact name do not need an EXTERNAL_MAP entry
LCFIT:
D:
BASE: surveys/des/lcfit_nml/des_5yer.nml
MASK: DESSIM
EXTERNAL_DIRS:
- $PIPPIN_OUTPUT/EXAMPLE_C11/2_LCFIT/DESFIT_SIM
- $PIPPIN_OUTPUT/EXAMPLE_G10/2_LCFIT/DESFIT_SIM
- $PIPPIN_OUTPUT/EXAMPLE/2_LCFIT/DESFIT_CCSIM
EXTERNAL_MAP:
# LCFIT_SIM: EXTERNAL_MASK
D_DESSIMBIAS5YRIA_C11: EXAMPLE_C11 # In this case we are matching to the pippin job name, as the LCFIT task name is shared between two EXTERNAL_DIRS
D_DESSIMBIAS5YRIA_G10: EXAMPLE_G10 # Same as C11
D_DESSIMBIAS5YRCC: DESFIT_CCSIM # In this case we match to the LCFIT task name, as the pippin job name (EXAMPLE) would match with the other EXTERNAL_DIRS
The EXTERNAL_MAP method should match with any pippin task which makes use of EXTERNAL_DIRS, and is only necessary if the EXTERNAL_DIRS task names do not match up.
If you have any issues using this new feature, check out the ref_des_5yr.yml file or flick me a message.
Pippin is essentially a wrapper around many different tasks. In this section, I'll try and explain how tasks are related to each other, and what each task is.
As a general note, most tasks have an OPTS where most details go. This is partially historical, but essentially properties
that Pippin uses to determine how to construct tasks (like MASK, classification mode, etc) are top level, and the Task itself gets passed everything
inside OPTS to use however it wants.
The DataPrep task is simple - it is mostly a pointer for Pippin towards an external directory that contains some photometry, to say we're going to make use of it. Normally this means data files, though you can also use it to point to simulations that have already been run to save yourself the hassle of rerunning them. The other thing the DataPrep task will do is run the new method of determining a viable initial guess for the peak time, which will be used by the light curve fitting task down the road. The full options available for the DataPrep task are:
DATAPREP:
SOMENAME:
OPTS:
# Location of the photometry files
RAW_DIR: $DES_ROOT/lcmerge/DESALL_forcePhoto_real_snana_fits
# Specify which types are confirmed Ia's, confirmed CC or unconfirmed. Used by ML down the line
TYPES:
IA: [101, 1]
NONIA: [20, 30, 120, 130]
# Blind the data. Defaults to True if SIM:True not set
BLIND: False
# Defaults to False. Important to set this flag if analysing a sim in the same way as data, as there
# are some subtle differences
SIM: False
# The method of estimating peak mjd values. Don't ask me what numbers mean what, ask Rick.
OPT_SETPKMJD: 16
The simulation task does exactly what you'd think it does. It invokes SNANA to run some similation as per your configuration. If something goes wrong, Pippin tries to dig through the log files to give you a useful error message, but sometimes this is difficult (i.e. the logs have been zipped up). With the current version of SNANA, each simulation can have at most one Ia component, and an arbitrary number of CC components. The specification for the simulation task config is as follows:
SIM:
SOMENAMEHERE:
# We specify the Ia component, so it must have IA in its name
IA_G10:
BASE: surveys/des/sims_ia/sn_ia_salt2_g10_des5yr.input # And then we specify the base input file which generates it.
# Now we can specify as many CC sims to mix in as we want
II_JONES:
BASE: surveys/des/sims_cc/sn_collection_jones.input
IAX:
BASE: surveys/des/sims_cc/sn_iax.input
DNDZ_ALLSCALE: 3.0 # Note you can add/overwrite keys like so for specific files
# This section will apply to all components of the sim
GLOBAL:
NGEN_UNIT: 1
RANSEED_REPEAT: 10 12345This task runs the SALT2 light curve fitting process on light curves from the simulation or DataPrep task. As above, if something goes wrong I try and give a good reason why, if you don't get a good reason, let me know. The task is specified like so:
LCFIT:
SOMENAMEHERE:
# MASK means only apply this light curve fitting on sims/Dataprep which have DES in the name
# You can also specify a list for this, and they will be applied as a logical or
MASK: DES
# The base nml file used
BASE: surveys/des/lcfit_nml/des.nml
# FITOPTS can be left out for nothing, pointed to a file, specified manually or a combination of the two
# Normally this would be a single entry like global.yml shown below, but you can also pass a list
# If you specify a FITOPT manually, make sure it has the / around the label
# And finally, if you specify a file, make sure its a yml dictionary that links a survey name to the correct
# fitopts. See the file below for an example
FITOPTS:
- surveys/global/lcfit_fitopts/global.yml
- "/custom_extra_fitopt/ REDSHIFT_FINAL_SHIFT 0.0001"
# We can optionally customise keys in the FITINP section
FITINP:
FILTLIST_FIT: 'gri'
# And do the same for the optional SNLCINP section
SNLCINP:
CUTWIN_SNRMAX: 3.0, 1.0E8
CUTWIN_NFILT_SNRMAX: 3.0, 99.
# Finally, options that go outside either of these sections just go in the generic OPTS
OPTS:
BATCH_INFO: sbatch $SBATCH_TEMPLATES/SBATCH_Midway2_1hr.TEMPLATE 10Within Pippin, there are many different classifiers implemented. Most classifiers need to be trained, and can then run in predict mode. All classifiers that require training can either be trained in the same yml file, or you can point to an external serialised instance of the trained class and use that. The general syntax for a classifier is:
CLASSIFICATION:
SOMELABEL:
CLASSIFIER: NameOfTheClass
MODE: train # or predict
MASK: optional_mask # Masks both sim and lcfit together, logical and
MASK_SIM: optional_sim_only_mask
MASK_FIT: optional_lcfit_only_mask
OPTS:
MODEL: file_or_label # only needed in predict mode, how to find the trained classifier
WHATREVER_THE: CLASSIFIER_NEEDS The SCONE classifier is a convolutional neural network-based classifier for supernova photometry. The model first creates "heatmaps" of flux values in wavelength-time space, then runs the neural network model on GPU (if available) to train or predict on these heatmaps. A successful run will produce predictions.csv, which shows the Ia probability of each SN. For debugging purposes, the model config (model_config.yml), Slurm job (job.slurm), log (output.log), and all the heatmaps (heatmaps/) can be found in the output directory. An example of how to define a SCONE classifier:
CLASSIFICATION:
SNN_TEST:
CLASSIFIER: SconeClassifier
MODE: predict # or train
# optional arguments
OPTS:
MODEL: /path/to/trained/model
GPU: False # defaults to True
CATEGORICAL: True # defaults to False (False = Ia vs. non-Ia classification)
SIM_FRACTION: 2 # defaults to 1, 1/SIM_FRACTION of the sims get used for classification (i.e. SIM_FRACTION=2 means 1/2 of the sims get used)
REMAKE_HEATMAPS: True # defaults to False (force remake heatmaps if prev run failed in an unusual way)
NUM_EPOCHS: 5 # defaults to 400, for training only
KCOR_FILE: /path/to/kcor/file # defaults to None
NUM_WAVELENGTH_BINS: 32 # defaults to 32, size of wavelength dimension of heatmap
NUM_MJD_BINS: 180 # defaults to 180, size of time dimension of heatmapThe SuperNNova classifier is a recurrent neural network that
operates on simulation photometry. It has three in vuilt variants - its normal (vanilla) mode, a Bayesian mode
and a Variational mode. After training, a model.pt can be found in the output directory,
which you can point to from a different yaml file. You can define a classifier like so:
CLASSIFICATION:
SNN_TEST:
CLASSIFIER: SuperNNovaClassifier
MODE: predict
GPU: True # Or False - determines which queue it gets sent into
CLEAN: True # Or false - determine if Pippin removes the processed folder to sae space
OPTS:
MODEL: SNN_TRAIN # Havent shown this defined. Or /somepath/to/model.pt
VARIANT: vanilla # or "variational" or "bayesian". Defaults to "vanilla"
REDSHIFT: True # Use redshift info when classifying. Defaults to True.
NORM: cosmo_quantile # How to normalise LCs. Other options are "perfilter", "cosmo", "global" or "cosmo_quantile".
CYCLIC: True # Defaults to True for vanilla and variational model
SEED: 0 # Sets random seed. Defaults to 0.
LIST_FILTERS: ['G', 'R', 'I', 'Z'] # What filters are present in the data, defaults to ['g', 'r', 'i', 'z']
SNTYPES: "/path/to/sntypes.txt" # Path to a file which lists the sn type mapping to be used. Example syntax for this can be found at https://github.com/LSSTDESC/plasticc_alerts/blob/main/Examples/plasticc_schema/elasticc_origmap.txt. Alternatively, yaml dictionaries can be used to specify each sn type individually.Pippin also allows for supernnova input yaml files to be passed, instead of having to define all of the options in the Pippin input yaml. This is done via:
OPTS:
DATA_YML: path/to/data_input.yml
CLASSIFICATION_YML: path/to/classification_input.ymlExample input yaml files can be found here, with the important variation that you must have:
raw_dir: RAW_DIR
dump_dir: DUMP_DIR
done_file: DONE_FILESo that Pippin can automatically replace these with the appropriate directories.
The SNIRF classifier is a random forest running off SALT2 summary
statistics. You can specify which features it gets to train on, which has a large impact on performance. After training,
there should be a model.pkl in the output directory. You can specify one like so:
CLASSIFICATION:
SNIRF_TEST:
CLASSIFIER: SnirfClassifier
MODE: predict
OPTS:
MODEL: SNIRF_TRAIN
FITOPT: some_label # Optional FITOPT to use. Match the label. Defaults to no FITOPT
FEATURES: x1 c zHD x1ERR cERR PKMJDERR # Columns to use. Defaults are shown. Check FITRES for options.
N_ESTIMATORS: 100 # Number of trees in forest
MIN_SAMPLES_SPLIT: 5 # Min number of samples to split a node on
MIN_SAMPLES_LEAF: 1 # Minimum number samples in leaf node
MAX_DEPTH: 0 # Max depth of tree. 0 means auto, which means as deep as it wants.Similar to SNIRF, NN trains on SALT2 summary statistics using a basic Nearest Neighbour algorithm from sklearn.
It will produce a model.pkl file in its output directory when trained. You can configure it as per SNIRF:
CLASSIFICATION:
NN_TEST:
CLASSIFIER: NearestNeighborPyClassifier
MODE: predict
OPTS:
MODEL: NN_TRAIN
FITOPT: some_label # Optional FITOPT to use. Match the label. Defaults to no FITOPT
FEATURES: zHD x1 c cERR x1ERR COV_x1_c COV_x1_x0 COV_c_x0 PKMJDERR # Columns to use. Defaults are shown.Sometimes you want to cheat, and if you have simulations, this is easy. The perfect classifier looks into the sims to get the actual type, and will then assign probabilities as per your configuration. This classifier has no training mode, only predict.
CLASSIFICATION:
PERFECT:
CLASSIFIER: PerfectClassifier
MODE: predict
OPTS:
PROB_IA: 1.0 # Probs to use for Ia events, default 1.0
PROB_CC: 0.0 # Probs to use for CC events, default 0.0To emulate a spectroscopically confirmed sample, or just to save time, we can assign every event a probability of 1.0 that it is a type Ia. As it just returns 1.0 for everything, it only has a predict mode
CLASSIFICATION:
UNITY:
CLASSIFIER: UnityClassifier
MODE: predictAnother useful debug test is to just take the SALT2 fit probability calculated from the chi2 fitting and use that as our probability. You'd hope that classifiers all improve on this. Again, this classifier only has a predict mode.
CLASSIFICATION:
FITPROBTEST:
CLASSIFIER: FitProbClassifier
MODE: predictThe aggregation task takes results from one or more classification tasks (that have been run in predict mode on the same dataset) and generates comparisons between the classifiers (their correlations, PR curves, ROC curves and their calibration plots). Additionally, it merges the results of the classifiers into a single csv file, mapping SNID to one column per classifier.
AGGREGATION:
SOMELABEL:
MASK: mask # Match sim AND classifier
MASK_SIM: mask # Match only sim
MASK_CLAS: mask # Match only classifier
RECALIBRATION: SIMNAME # Optional, use this simulation to recalibrate probabilities. Default no recal.
# Optional, changes the probability column name of each classification task listed into the given probability column name.
# Note that this will crash if the same classification task is given multiple probability column names.
# Mostly used when you have multiple photometrically classified samples
MERGE_CLASSIFIERS:
PROB_COLUMN_NAME: [CLASS_TASK_1, CLASS_TASK_2, ...]
OPTS:
PLOT: True # Default True, make plots
PLOT_ALL: False # Default False. Ie if RANSEED_CHANGE gives you 100 sims, make 100 set of plots.The merging task will take the outputs of the aggregation task, and put the probabilities from each classifier into the light curve fit results (FITRES files) using SNID.
MERGE:
label:
MASK: mask # partial match on all sim, fit and agg
MASK_SIM: mask # partial match on sim
MASK_FIT: mask # partial match on lcfit
MASK_AGG: mask # partial match on aggregation taskWith all the probability goodness now in the FITRES files, we can move onto calculating bias corrections. For spec-confirmed surveys, you only need a Ia sample for bias corrections. For surveys with contamination, you will also need a CC only simulation/lcfit result. For each survey being used (as we would often combine lowz and highz surveys), you can specify inputs like below.
Note that I expect this task to have the most teething issues, especially when we jump into the MUOPTS.
BIASCOR:
LABEL:
# The base input file to utilise
BASE: surveys/des/bbc/bbc.input
# The names of the lcfits_data/simulations going in. List format please. Note LcfitLabel_SimLabel format
DATA: [DESFIT_DESSIM, LOWZFIT_LOWZSIM]
# Input Ia bias correction simulations to be concatenated
SIMFILE_BIASCOR: [DESFIT_DESBIASCOR, LOWZFIT_LOWZBIASCOR]
# For surveys that have contamination, add in the cc only simulation under CCPRIOR
SIMFILE_CCPRIOR: DESFIT_DESSIMBIAS5YRCC
# Which classifier to use. Column name in FITRES will be determined from this property.
# In the case of multiple classifiers this can either be
# 1. A list of classifiers which map to the same probability column name (as defined by MERGE_CLASSIFIERS in the AGGREGATION stage)
# 2. A probability column name (as defined by MERGE_CLASSIFIERS in the AGGREGATION stage)
# Note that this will crash if the specified classifiers do not map to the same probability column.
CLASSIFIER: UNITY
# Default False. If multiple sims (RANSEED_CHANGE), make one or all Hubble plots.
MAKE_ALL_HUBBLE: False
# Defaults to False. Will load in the recalibrated probabilities, and crash and burn if they dont exist.
USE_RECALIBRATED: True
# Defaults to True. If set to True, will rerun biascor twice, removing any SNID that got dropped in any FITOPT/MUOPT
CONSISTENT_SAMPLE: False
# We can also specify muopts to add in systematics. They share the structure of the main biascor definition
# You can have multiple, use a dict structure, with the muopt name being the key
MUOPTS:
C11:
SIMFILE_BIASCOR: [D_DESBIASSYS_C11, L_LOWZBIASSYS_C11]
SCALE: 0.5 # Defaults to 1.0 scale, used by CREATE_COV to determine covariance matrix contribution
# Generic OPTS that can modify the base file and overwrite properties
OTPS:
BATCH_INFO: sbatch $SBATCH_TEMPLATES/SBATCH_Midway2_1hr.TEMPLATE 10For those that generate large simulations and want to cut them up into little pieces, you want the NSPLITRAN syntax.
The configuration below will take the inputs and divide them into 10 samples, which will then propagate to 10 CosmoMC runs
if you have a CosmoMC task defined.
BIASCOR:
LABEL:
BASE: surveys/des/bbc/bbc_3yr.input
DATA: [D_DES_G10]
SIMFILE_BIASCOR: [D_DESSIMBIAS3YRIA_G10]
PROB_COLUMN_NAME: some_column_name # optional instead of CLASSIFIER
OPTS:
NSPLITRAN: 10Assuming the biascor task hasn't died, its time to prep for CosmoMC. To do this, we invoke a script from Dan originally (I think) that essentially creates all the input files and structure needed by CosmoMC. It provides a way of scaling systematics, and determining which covariance options to run with.
CREATE_COV:
SOMELABEL:
MASK: some_biascor_task
OPTS:
INI_DIR: /path/to/your/own/dir/of/cosmomc/templates # Defaults to cosmomc_templates, which you can exploit using DATA_DIRS
SYS_SCALE: surveys/global/lcfit_fitopts/global.yml # Location of systematic scaling file, same as the FITOPTS file.
SINGULAR_BLIND: False # Defaults to False, whether different contours will have different shifts applied
BINNED: True # Whether to bin the SN or not for the covariance matrx. Defaults to True
REBINNED_X1: 2 # Rebin x1 into 2 bins
REBINNED_C: 4 # Rebin c into 4 bins
SUBTRACT_VPEC: False # Subtract VPEC contribution to MUERR if True. Used when BINNED: False
FITOPT_SCALES: # Optional
FITOPT_LABEL: some_scale # Note this is a partial match, ie SALT2: 1.0 would apply to all SALT2 cal fitopts
MUOPT_SCALES:
MUOPT_LABEL: some_scale # This is NOT a partial match, must be exact
COVOPTS: # Optional, and you'll always get an 'ALL' covopt. List format please
- "[NOSYS] [=DEFAULT,=DEFAULT]" # This syntax is explained belowIf you don't specify SYS_SCALE, Pippin will search the LCFIT tasks from the BIASCOR dependency and if all LCFIT tasks
have the same fitopt file, it will use that.
The COVOPTS section is a bit odd. In the square brackets first, we have the label that will be assigned and used
in the plotting output later. The next set of square backets is a two-tuple, and it applies to [fitopts,muopts] in
that order. For example, to get four contours out of CosmoMC corresponding to all uncertainty, statistics only,
statistics + calibration uncertainty, and fitopts + C11 uncertainty, we could set:
COVOPTS:
- "[NOSYS] [=DEFAULT,=DEFAULT]"
- "[CALIBRATION] [+cal,=DEFAULT]"
- "[SCATTER] [=DEFAULT,=C11]"CosmoFit is a generic cosmological fitting task, which allows you to choose between different fitters. The syntax is very simple:
COSMOFIT:
COSMOMC:
SOMELABEL:
# CosmoMC options
WFIT:
SOMEOTHERLABEL:
# WFit optionsLaunching CosmoMC is hopefully fairly simple. There are a list of provided configurations under the cosmomc_templates
directory (inside data_files), and the main job of the user is to pick which one they want.
COSMOFIT:
COSMOMC:
SOMELABEL:
MASK_CREATE_COV: mask # partial match
OPTS:
INI: sn_cmb_omw # should match the filename of an ini file
NUM_WALKERS: 8 # Optional, defaults to eight.
# Optional, covopts from CREATE_COV step to run against. If blank, you get them all. Exact matching.
COVOPTS: [ALL, NOSYS]Launching WFit simply requires providing the command line options you want to use for each fit.
COSMOFIT:
WFIT:
SOMELABEL:
MASK: mask # partial match
OPTS:
BATCH_INFO: sbatch path/to/SBATCH.TEMPLATE 10 # Last number is the number of cores
WFITOPT_GLOBAL: "-hsteps 61 -wsteps 101 -omsteps 81" # Optional, will apply these options to all fits"
WFITOPTS:
- /om_pri/ -ompri 0.31 -dompri 0.01 # At least one option is required. The name in the /'s is a human readable label
- /cmb_pri/ -cmb_sim -sigma_Rcmb 0.007 # Optionally include as many other fitopts as you want.
The final step in the Pippin pipeline is the Analyse task. It creates a final output directory, moves relevant files into it,
and generates extra plots. It will save out compressed CosmoMC chains and the plotting scripts (so you can download
the entire directory and customise it without worrying about pointing to external files), it will copy in Hubble diagrams,
and - depending on if you've told it to, will make histogram comparison plots between data and sim. Oh and also
redshift evolution plots. The scripts which copy/compress/rename external files into the analyse directory are generally
named parse_*.py. So parse_cosmomc.py is the script which finds, reads and compresses the MCMC chains from CosmoMC into
the output directory. Then plot_cosmomc.py reads those compressed files to make the plots.
Cosmology contours will be blinded when made by looking at the BLIND flag set on the data. For data, this defaults to True.
Note that all the plotting scripts work the same way - Analyse generates a small yaml file pointing to all the
resources called input.yml, and each script uses the same file to make different plots. It is thus super easy to add your own
plotting code scripts, and you can specify arbitrary code to execute using the ADDITIONAL_SCRIPTS keyword in opts.
Just make sure your code takes input.yml as an argument. As an example, to rerun the CosmoMC plots, you'd simply have to
run python plot_cosmomc.py input.yml.
ANALYSE:
SOMELABEL:
MASK_COSMOFIT: mask # partial match
MASK_BIASCOR: mask # partial match
MASK_LCFIT: [D_DESSIM, D_DATADES] # Creates histograms and efficiency based off the input LCFIT_SIMNAME matches. Optional
OPTS:
COVOPTS: [ALL, NOSYS] # Optional. Covopts to match when making contours. Single or list. Exact match.
SHIFT: False # Defualt False. Shift all the contours on top of each other
PRIOR: 0.01 # Default to None. Optional normal prior around Om=0.3 to apply for sims if wanted.
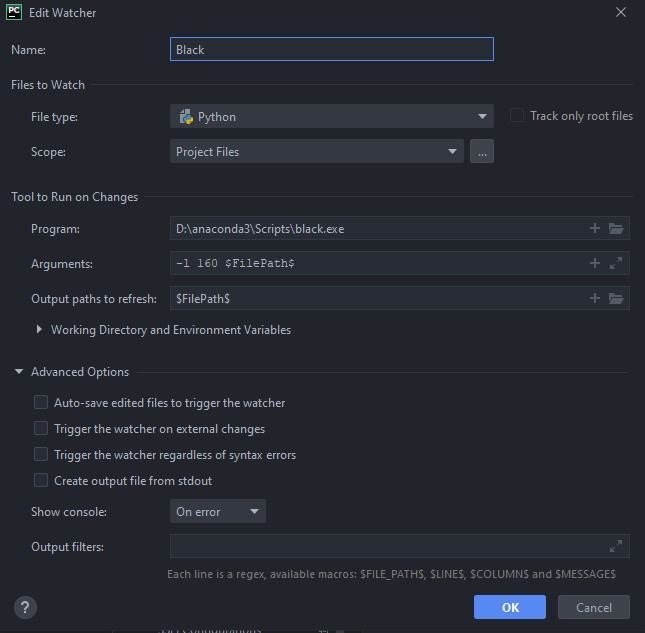
ADDITIONAL_SCRIPTS: /somepath/to/your/script.py # Should take the input.yml as an argumentPlease, for the love of god, don't code this up in vim/emacs on a terminal connection. Use a proper IDE (I recommend PyCharm or VSCode), and install the Black extensiion! I have Black set up in PyCharm as a file watcher, and all python files, on save, are automatically formatted. Use 160 characters a linewidth. Here is the Black file watcher config:
If everyone does this, then all files should remain consistent across different users.
Click for the gory details
To ensure we don't break things when pushing out new code, the tests directory contains a set of tests progressively increasing in pipeline complexity, designed to ensure that existing config files act consistently regardless of code changes. Any failure in the tests means a break in backwards compatibility and should be discussed before being incorporated into a release.
To run the tests, in the top level directory, simply run:
pytest -v .
Click for the gory details
Alright there, you want to add a new task to Pippin? Great. Here's what you've got to do:
- Create an implementation of the
Taskclass, can keep it empty for now. - Figure out where it goes - in
manager.pyat the top you can see the current stages in Pippin. You'll probably need to figure out where it should go. Once you have figured it out, import the task and slot it in. - Back in your new class that extends Task, you'll notice you have a few methods to implement:
-
_run(): Kick the task off, report True or False for successful kicking off. To help with determining the hash and whether the task shoudl run, there are a few handy functions:_check_regenerate,get_hash_from_string,save_hash,get_hash_from_files,get_old_hash. See, for example, the Analyse task for an example on how I use these. -
_check_completion(squeue): Check to see if the task (whether its being rerun or not) is done. Normally I do this by checking for a done file, which contains either SUCCESS or FAILURE. For example, if submitting a script to a queuing system, I might have this after the primary command:if [ $? -eq 0 ]; then echo SUCCESS > {done_file} else echo FAILURE > {done_file} fi
This allows me to easily see if a job failed or passed. On failure, I then generally recommend looking through the task logs and trying to figure out what went wrong, so you can present a useful message to your user. To then show that error, or ANY MESSAGE TO THE USER, use the provided logger:
self.logger.error("The task failed because of this reason").This method should return either a) Task.FINISHED_FAILURE, Task.FINISHED_SUCCESS, or alternatively the number of jobs still in the queue, which you could figure out because I pass in all jobs the user has active in the variable squeue (which can sometimes be None).
-
get_tasks(task_config, prior_tasks, output_dir, stage_num, prefix, global_config): From the given inputs, determine what tasks should be created, and create them, and then return them in a list. For context, here is the code I use to determine what simulation tasks to create:@staticmethod def get_tasks(config, prior_tasks, base_output_dir, stage_number, prefix, global_config): tasks = [] for sim_name in config.get("SIM", []): sim_output_dir = f"{base_output_dir}/{stage_number}_SIM/{sim_name}" s = SNANASimulation(sim_name, sim_output_dir, f"{prefix}_{sim_name}", config["SIM"][sim_name], global_config) Task.logger.debug(f"Creating simulation task {sim_name} with {s.num_jobs} jobs, output to {sim_output_dir}") tasks.append(s) return tasks
-
Click for the gory details
Alright, so what if we're not after a brand new task, but just adding another classifier. Well, its easier to do, and I recommend looking at
nearest_neighbor_python.py for something to copy from. You'll see we have the parent Classifier class, I write out the slurm script that
would be used, and then define the train and predict method (which both invoke a general classify function in different ways, you can do this
however you want.)
You'll also notice a very simply _check_completion method, and a get_requirmenets method. The latter returns a two-tuple of booleans, indicating
whether the classifier needs photometry and light curve fitting results respectively. For the NearestNeighbour code, it classifies based
only on SALT2 features, so I return (False, True).
Finally, you'll need to add your classifier into the ClassifierFactory in classifiers/factory.py, so that I can link a class name
in the YAML configuration to your actual class. Yeah yeah, I could use reflection or dynamic module scanning or similar, but I've had issues getting
the behaviour consistent across systems and conda environments, so we're doing it the hard way.