ICR142 validation in bcbio
Support running an ICR142 validation using bcbio
http://f1000research.com/articles/5-386/v1
Running validation
This repository contains a full set of configuration files and BED/VCF validation sets to run an analysis with bcbio:
-
Obtain the ICR142 fastq files, which require applying for access. Move these to
bcbiorun/input/fastqs -
Run the analysis using an installed version of bcbio. This can run on a single machine using multiple cores or distributed on a cluster:
cd bcbiorun/work bcbio_nextgen.py ../config/icr142.yaml -n 16 -
Summarize and plot the results:
cd ../summarize bcbio_python ../../scripts/combine_samples.py bcbio_python ../../scripts/bcbio_validation_plot.py icr142-summary.csv
Results
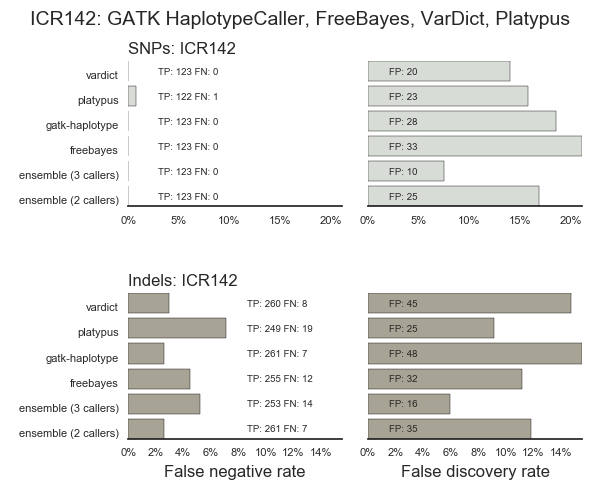
Validation using bwa-mem and 3 variant callers (GATK HaplotypeCaller, FreeBayes and VarDict), including ensemble regions with calls in 2 of our 3 or 3 out of 3 callers. The majority of false positives are present in at least 2 callers, and many in all 3:
Truth set preparation
We prepared the truth set and analysis regions using the truth set calls from
Supplemental table 1:
scripts/icr_to_vcf.py created the VCF and BED files contained in the
repository from the original table and a list of variants found to be homozygous
(both in bcbiorun/input). The initial truth table does not have information
about whether exepcted variants are homozygous or heterozygous so we ran an
intial validation with everything heterozygous, then used
scripts/find_hethomerrors.py to find those variants that are likely homozygous
to reprepare the final truth set.