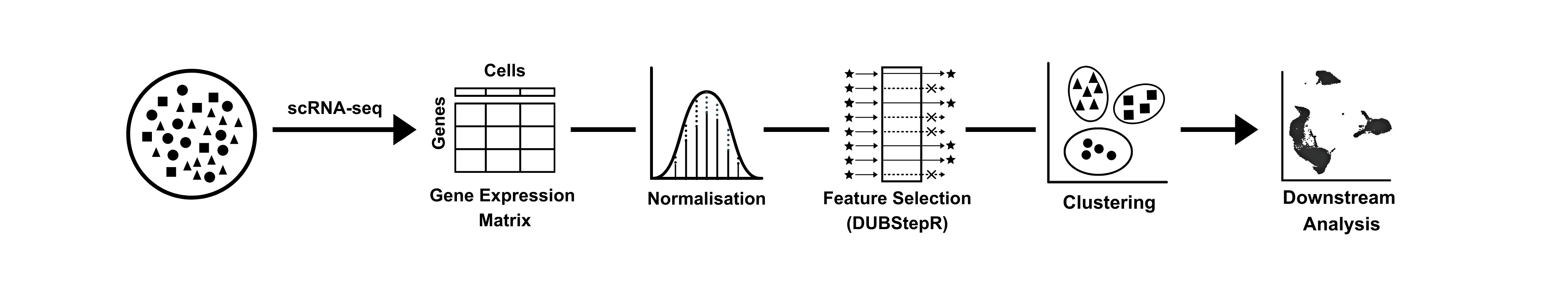
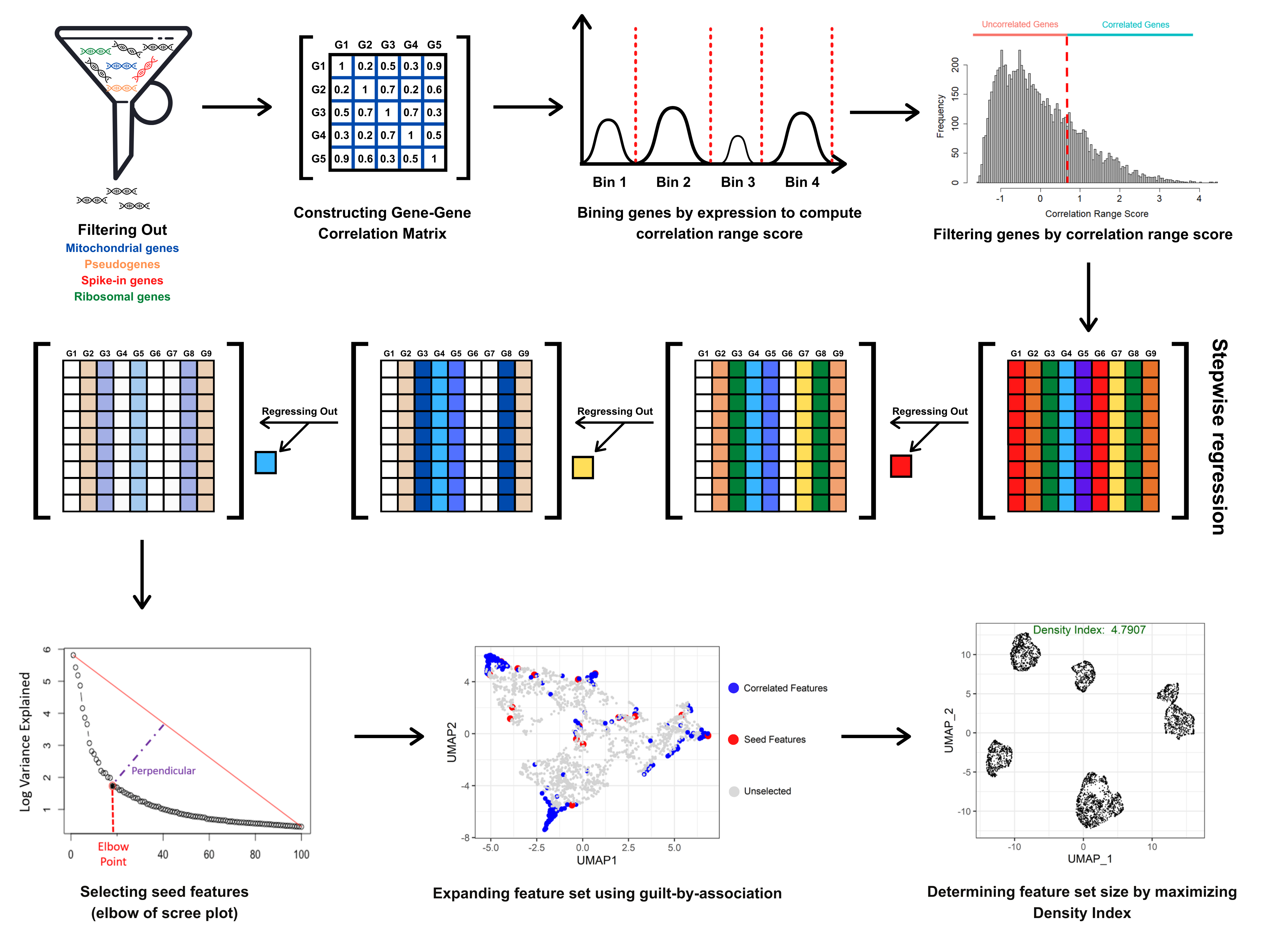
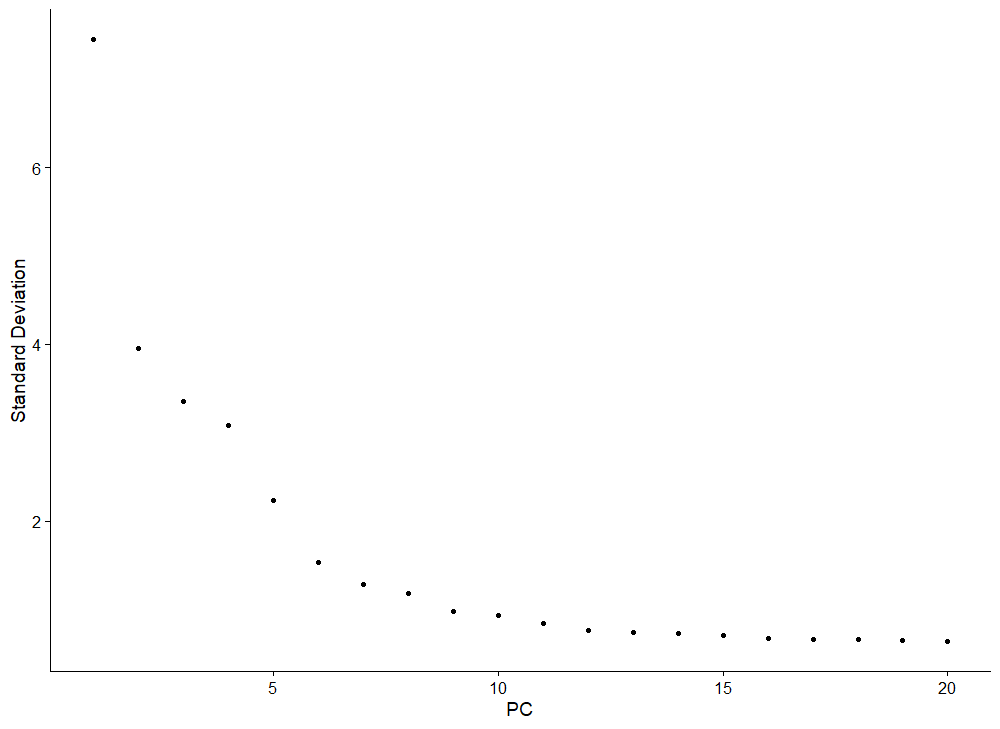DUBStepR (Determining the Underlying Basis using Step-wise Regression) is a feature selection algorithm for cell type identification in single-cell RNA-sequencing data. Please refer to https://github.com/prabhakarlab/DUBStepR for the latest version of DUBStepR.
Feature selection, i.e. determining the optimal subset of genes to cluster cells into cell types, is a critical step in the unsupervised clustering of scRNA-seq data.
DUBStepR is based on the intuition that cell-type-specific marker genes tend to be well correlated with each other, i.e. they typically have strong positive and negative correlations with other marker genes. After filtering genes based on a correlation range score, DUBStepR exploits structure in the gene-gene correlation matrix to prioritize genes as features for clustering.
Version 1.0 released on 10 December 2019.
DUBStepR requires your R version to be >= 3.5.0. Once you've ensured that, you can install DUBStepR from GitHub using the following commands:
install.packages("devtools")
devtools::install_github("prabhakarlab/DUBStepR")The installation should take ~ 30 seconds, not including dependencies.
After installation, load DUBStepR using the following command:
library(DUBStepR)For users new to single-cell RNA sequencing data analysis, we recommend using DUBStepR with the Seurat (Satija, R. et al. Nature Biotechnol. 33.5(2015):495.) package to analyze single-cell RNA seq data. Below is a tutorial using the Seurat package.
Seurat can be installed and loaded into your R environment using the following commands:
install.packages("Seurat")
library(Seurat)
library(dplyr)Here, we use a publicly available PBMC dataset generated by 10X Genomics. Here's a link to the dataset. We use the Feature / cell matrix HDF5 (filtered) file.
Locate the file in your working directory and load the data in to your Seurat object in the following manner:
seuratObj <- CreateSeuratObject(counts = Read10X_h5("5k_pbmc_v3_filtered_feature_bc_matrix.h5"), assay = "RNA", project = "10k_PBMC")
seuratObjAn object of class Seurat
33538 features across 5025 samples within 1 assay
Active assay: RNA (33538 features)
For DUBStepR, we recommend log-normalizing your 10X data. That can be performed in Seurat using the following command:
seuratObj <- NormalizeData(object = seuratObj, normalization.method = "LogNormalize")DUBStepR can be inserted into the Seurat workflow at this stage, and we recommend that be done in the following manner:
dubstepR.out <- DUBStepR(input.data = seuratObj@assays$RNA@data, min.cells = 0.05*ncol(seuratObj), optimise.features = T, k = 10, num.pcs = 20, error = 0)
seuratObj@assays$RNA@var.features <- dubstepR.out$optimal.feature.genes[1] "Expression Filtering - Done"
[1] "Mitochondrial, Ribosomal and Pseudo Genes Filtering - Done"
[1] "Computing GGC.."
[1] "done."
[1] "Running Stepwise Regression"
|=======================================================================================================================================|100%
[1] "Adding correlated features"
|=======================================================================================================================================|100%
[1] "Determining optimal feature set"
|=============================================================================================================================| 100%
This step could take upto 12 minutes on a normal desktop computer.
Following Seurat's recommendations, we scale the gene expression data and run Principal Component Analysis (PCA). We then visualize the standard deviation of PCs using an elbow plot and select the number of PCs we think is sufficient to explain the variance in the dataset.
seuratObj <- ScaleData(seuratObj, features = rownames(seuratObj))
seuratObj <- RunPCA(seuratObj, features = VariableFeatures(object = seuratObj))
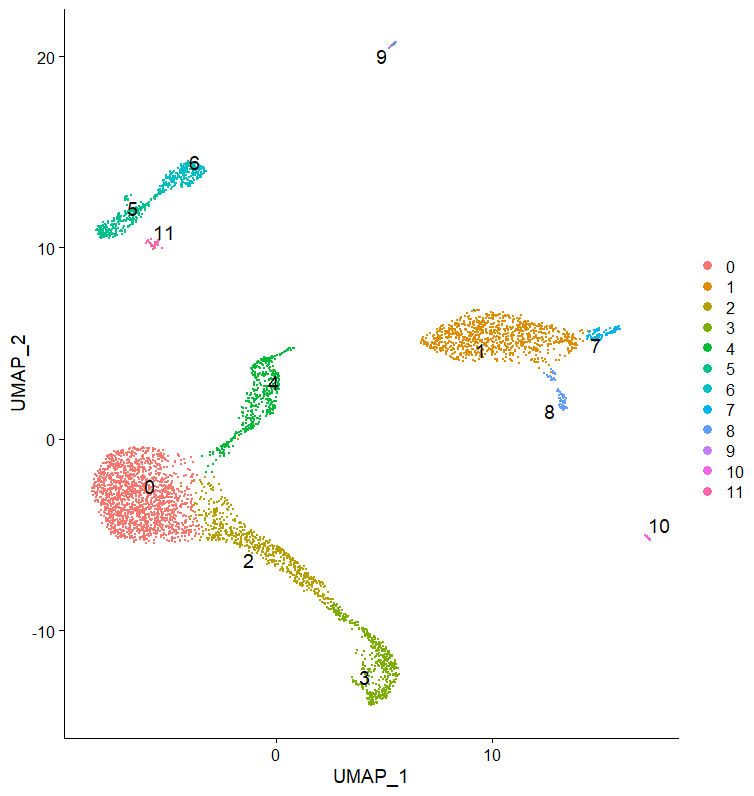
ElbowPlot(seuratObj)We select 10 PCs for clustering, and visualize the cells in a 2D UMAP.
seuratObj <- FindNeighbors(seuratObj, reduction = "pca", dims = 1:10)
seuratObj <- FindClusters(seuratObj, resolution = 0.4)
seuratObj <- RunUMAP(seuratObj, dims = 1:10, n.components = 2, seed.use = 2019)
DimPlot(seuratObj, reduction = "umap", label = TRUE, pt.size = 0.5, repel = T, label.size = 5)
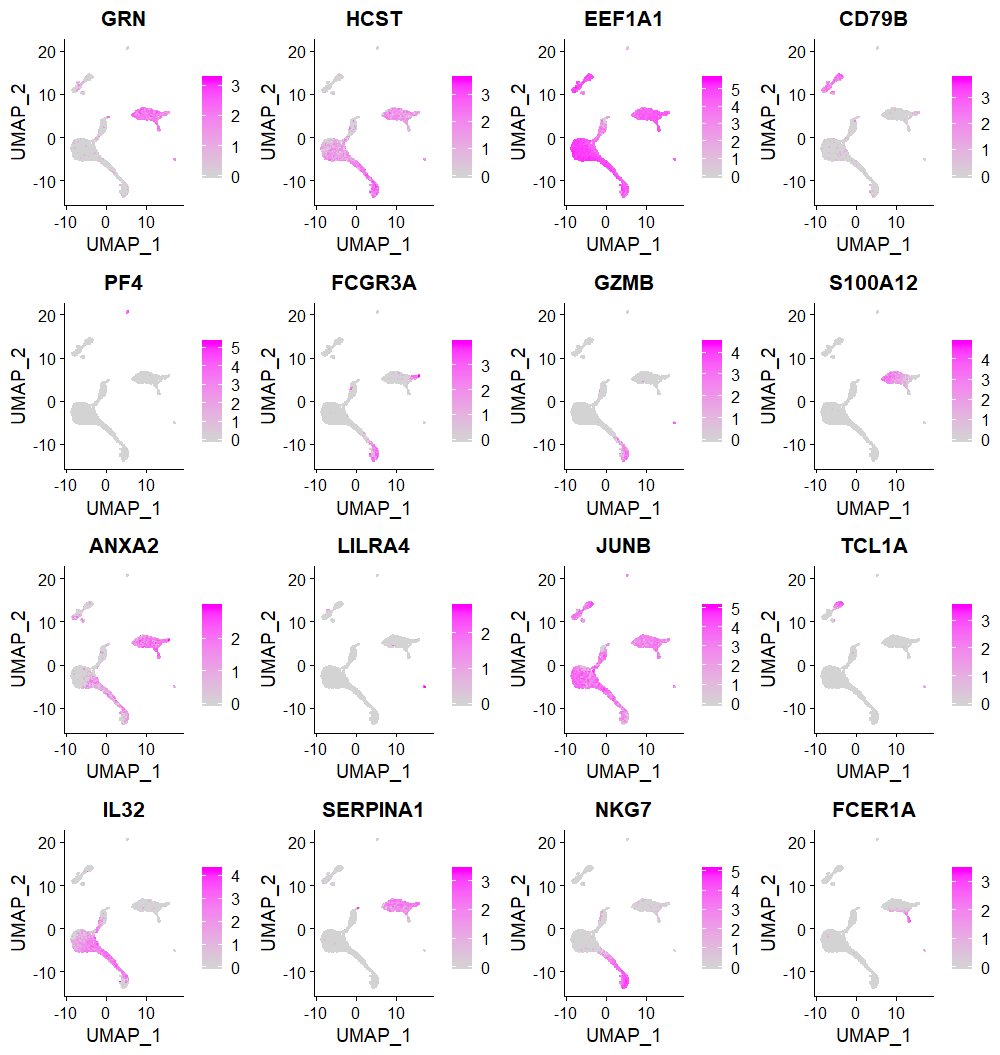
We select the first few feature genes selected by DUBStepR to show cell type specific expression
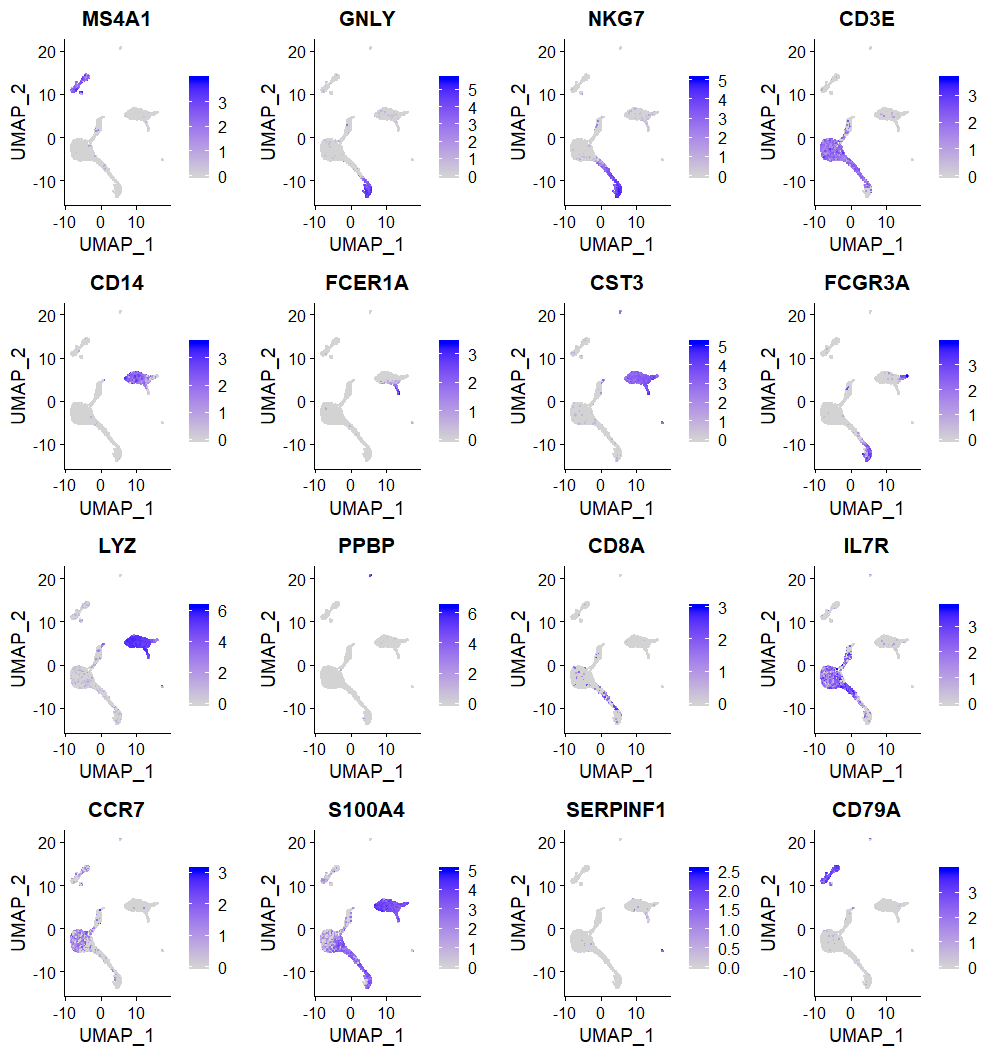
FeaturePlot(seuratObj, features = VariableFeatures(object = seuratObj)[1:16], cols = c("lightgrey", "magenta"))Using known marker genes, we show cell type specific regions of the UMAP
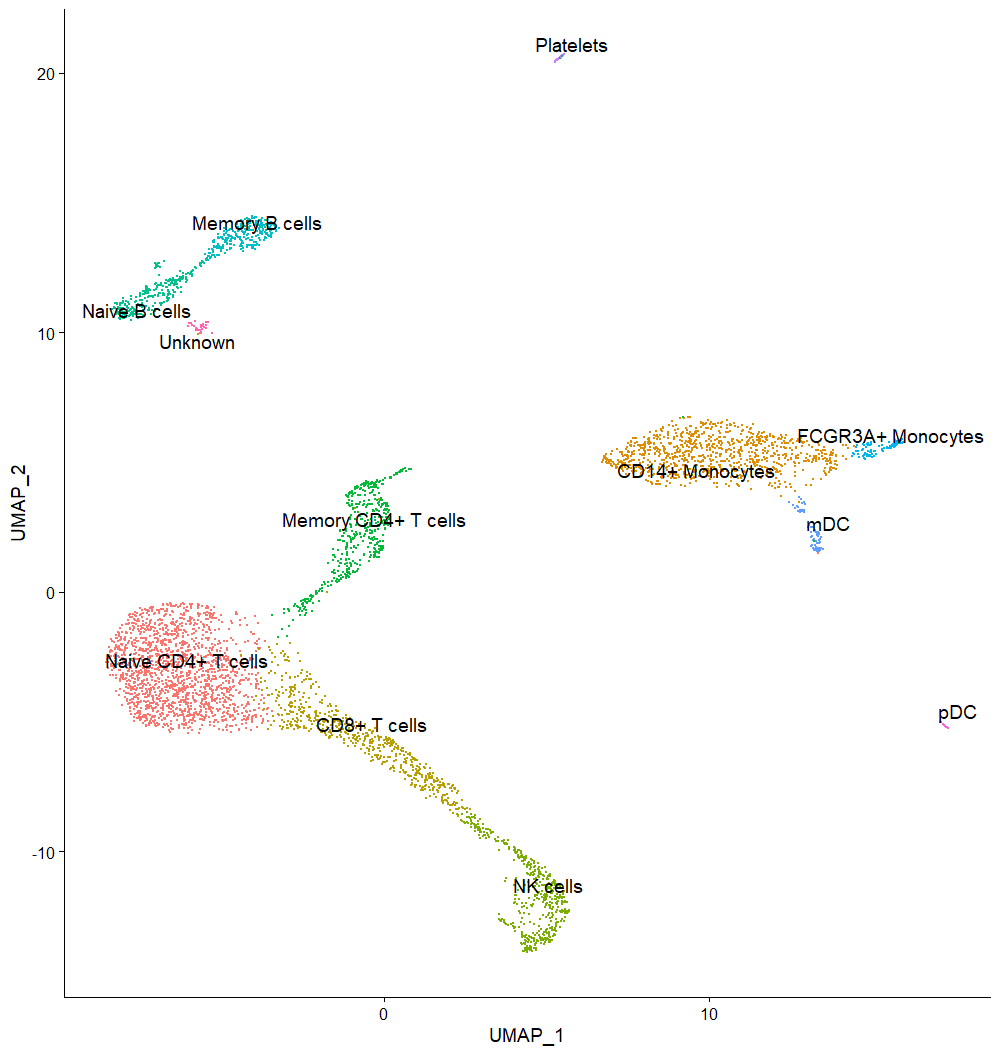
FeaturePlot(seuratObj, features = c("MS4A1", "GNLY", "NKG7", "CD3E", "CD14", "FCER1A", "CST3", "FCGR3A", "LYZ", "PPBP", "CD8A", "IL7R", "CCR7", "S100A4", "SERPINF1", "CD79A"))Annotating clusters using gene expression
cell.types <- c("Naive CD4+ T cells", "CD14+ Monocytes", "CD8+ T cells", "NK cells", "Memory CD4+ T cells", "Naive B cells", "Memory B cells", "FCGR3A+ Monocytes", "mDC", "Platelets", "pDC", "Unknown")
names(cell.types) <- levels(seuratObj)
seuratObj <- RenameIdents(seuratObj, cell.types)
DimPlot(seuratObj, reduction = "umap", label = TRUE, pt.size = 0.5, repel = T, label.size = 5) + NoLegend()