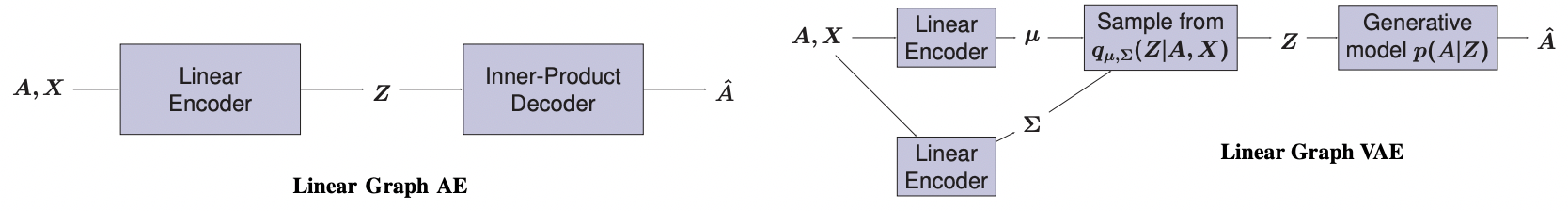
This repository provides Python code to reproduce experiments from the article Keep It Simple: Graph Autoencoders Without Graph Convolutional Networks presented at the NeurIPS 2019 Workshop on Graph Representation Learning.
Update: an extended conference version of this article is now available here: Simple and Effective Graph Autoencoders with One-Hop Linear Models.
We release Tensorflow implementations of the following two graph embedding models from the paper:
- Linear Graph Autoencoders
- Linear Graph Variational Autoencoders
together with standard Graph Autoencoders (AE) and Graph Variational Autoencoders (VAE) models (with 2-layer or 3-layer Graph Convolutional Networks encoders) from Kipf and Welling (2016).
We evaluate all models on the link prediction and node clustering tasks introduced in the paper. We provide the Cora, Citeseer and Pubmed datasets in the data folder, and refer to section 4 of the paper for direct link to the additional datasets used in our experiments.
Our code builds upon Thomas Kipf's original Tensorflow implementation of standard Graph AE/VAE.
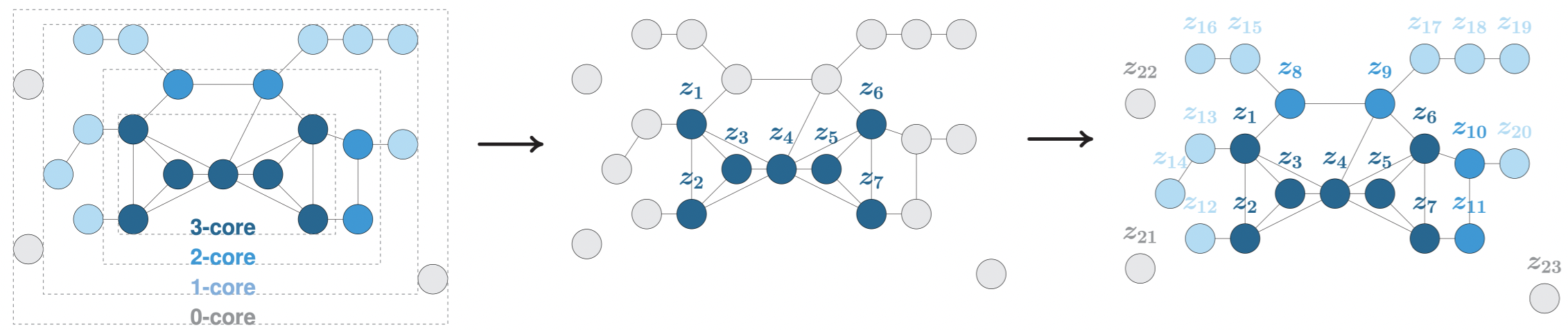
Graph AE and VAE training has a quadratic time complexity w.r.t. the number of nodes in the graph. In order to scale graph autoencoders to large graphs with millions of nodes and egdes, we also provide an implementation of our framework from the article A Degeneracy Framework for Scalable Graph Autoencoders, published in the proceedings of the 28th International Joint Conference on Artificial Intelligence (IJCAI 2019).
In this paper, we propose to train the graph AE/VAE only from a dense subset of nodes, namely the k-core or k-degenerate subgraph, instead of using the entire graph. Then, we propagate embedding representations to the remaining nodes using faster heuristics. We argue in the IJCAI paper that such strategy can significantly improve speed and scalability while preserving good performance.
python setup.py installRequirements: tensorflow (1.X), networkx, numpy, scikit-learn, scipy
cd linear_gae
python train.py --model=gcn_vae --dataset=cora --task=link_prediction
python train.py --model=linear_vae --dataset=cora --task=link_predictionThe above commands will train a standard Graph VAE with 2-layer GCN encoders (line 2) and a Linear Graph VAE (line 3) on Cora dataset and will evaluate embeddings on the Link Prediction task, with all parameters set to default values.
python train.py --model=gcn_vae --dataset=cora --task=link_prediction --kcore=True --k=2
python train.py --model=gcn_vae --dataset=cora --task=link_prediction --kcore=True --k=3
python train.py --model=gcn_vae --dataset=cora --task=link_prediction --kcore=True --k=4By adding --kcore=True, the model will only be trained on the k-core subgraph instead of using the entire graph. Here, k is a parameter (from 0 to the maximal core number of the graph) to specify using the --k flag.
| Parameter | Type | Description | Default Value |
|---|---|---|---|
model |
string | Name of the model, among: - gcn_ae: Graph AE from Kipf and Welling (2016), with 2-layer GCN encoder and inner product decoder- gcn_vae: Graph VAE from Kipf and Welling (2016), with Gaussian distributions, 2-layer GCN encoders for mu and sigma, and inner product decoder - linear_ae: Linear Graph AE, as introduced in section 3 of NeurIPS workshop paper, with linear encoder, and inner product decoder - linear_vae: Linear Graph VAE, as introduced in section 3 of NeurIPS workshop paper, with Gaussian distributions, linear encoders for mu and sigma, and inner product decoder - deep_gcn_ae: Deeper version of Graph AE, with 3-layer GCN encoder, and inner product decoder - deep_gcn_vae: Deeper version of Graph VAE, with Gaussian distributions, 3-layer GCN encoders for mu and sigma, and inner product decoder |
gcn_ae |
dataset |
string | Name of the dataset, among: - cora: scientific publications citation network - citeseer: scientific publications citation network - pubmed: scientific publications citation network We provide the preprocessed versions, coming from the tkipf/gae repository. Please check the LINQS website for raw data You can specify any additional graph dataset, in edgelist format, by editing input_data.py |
cora |
task |
string | Name of the Machine Learning evaluation task, among: - link_prediction: Link Prediction - node_clustering: Node Clustering See section 4 and supplementary material of NeurIPS 2019 workshop paper for details about tasks |
link_prediction |
dropout |
float | Dropout rate | 0. |
epoch |
int | Number of epochs in model training | 200 |
features |
boolean | Whether to include node features in encoder | False |
learning_rate |
float | Initial learning rate (with Adam optimizer) | 0.01 |
hidden |
int | Number of units in GCN encoder hidden layer(s) | 32 |
dimension |
int | Dimension of encoder output, i.e. embedding dimension | 16 |
kcore |
boolean | Whether to run k-core decomposition and use the degeneracy framework from IJCAI paper. If False, the AE/VAE will be trained on the entire graph |
False |
k |
int | Which k-core to use. Higher k => smaller graphs and faster (but maybe less accurate) training | 2 |
nb_run |
integer | Number of model runs + tests | 1 |
prop_val |
float | Proportion of edges in validation set (for Link Prediction) | 5. |
prop_test |
float | Proportion of edges in test set (for Link Prediction) | 10. |
validation |
boolean | Whether to report validation results at each epoch (for Link Prediction) | False |
verbose |
boolean | Whether to print full comments details | True |
Cora
python train.py --dataset=cora --model=linear_ae --task=link_prediction --epochs=200 --learning_rate=0.01 --dimension=16 --nb_run=5
python train.py --dataset=cora --model=linear_vae --task=link_prediction --epochs=200 --learning_rate=0.01 --dimension=16 --nb_run=5
python train.py --dataset=cora --model=gcn_ae --task=link_prediction --epochs=200 --learning_rate=0.01 --hidden=32 --dimension=16 --nb_run=5
python train.py --dataset=cora --model=gcn_vae --task=link_prediction --epochs=200 --learning_rate=0.01 --hidden=32 --dimension=16 --nb_run=5
python train.py --dataset=cora --model=deep_gcn_ae --task=link_prediction --epochs=200 --learning_rate=0.01 --hidden=32 --dimension=16 --nb_run=5
python train.py --dataset=cora --model=deep_gcn_vae --task=link_prediction --epochs=200 --learning_rate=0.01 --hidden=32 --dimension=16 --nb_run=5Cora - with features
python train.py --dataset=cora --features=True --model=linear_ae --task=link_prediction --epochs=200 --learning_rate=0.01 --dimension=16 --nb_run=5
python train.py --dataset=cora --features=True --model=linear_vae --task=link_prediction --epochs=200 --learning_rate=0.01 --dimension=16 --nb_run=5
python train.py --dataset=cora --features=True --model=gcn_ae --task=link_prediction --epochs=200 --learning_rate=0.01 --hidden=32 --dimension=16 --nb_run=5
python train.py --dataset=cora --features=True --model=gcn_vae --task=link_prediction --epochs=200 --learning_rate=0.01 --hidden=32 --dimension=16 --nb_run=5
python train.py --dataset=cora --features=True --model=deep_gcn_ae --task=link_prediction --epochs=200 --learning_rate=0.01 --hidden=32 --dimension=16 --nb_run=5
python train.py --dataset=cora --features=True --model=deep_gcn_vae --task=link_prediction --epochs=200 --learning_rate=0.01 --hidden=32 --dimension=16 --nb_run=5Citeseer
python train.py --dataset=citeseer --model=linear_ae --task=link_prediction --epochs=200 --learning_rate=0.01 --dimension=16 --nb_run=5
python train.py --dataset=citeseer --model=linear_vae --task=link_prediction --epochs=200 --learning_rate=0.01 --dimension=16 --nb_run=5
python train.py --dataset=citeseer --model=gcn_ae --task=link_prediction --epochs=200 --learning_rate=0.01 --hidden=32 --dimension=16 --nb_run=5
python train.py --dataset=citeseer --model=gcn_vae --task=link_prediction --epochs=200 --learning_rate=0.01 --hidden=32 --dimension=16 --nb_run=5
python train.py --dataset=citeseer --model=deep_gcn_ae --task=link_prediction --epochs=200 --learning_rate=0.01 --hidden=32 --dimension=16 --nb_run=5
python train.py --dataset=citeseer --model=deep_gcn_vae --task=link_prediction --epochs=200 --learning_rate=0.01 --hidden=32 --dimension=16 --nb_run=5Citeseer - with features
python train.py --dataset=citeseer --features=True --model=linear_ae --task=link_prediction --epochs=200 --learning_rate=0.01 --dimension=16 --nb_run=5
python train.py --dataset=citeseer --features=True --model=linear_vae --task=link_prediction --epochs=200 --learning_rate=0.01 --dimension=16 --nb_run=5
python train.py --dataset=citeseer --features=True --model=gcn_ae --task=link_prediction --epochs=200 --learning_rate=0.01 --hidden=32 --dimension=16 --nb_run=5
python train.py --dataset=citeseer --features=True --model=gcn_vae --task=link_prediction --epochs=200 --learning_rate=0.01 --hidden=32 --dimension=16 --nb_run=5
python train.py --dataset=citeseer --features=True --model=deep_gcn_ae --task=link_prediction --epochs=200 --learning_rate=0.01 --hidden=32 --dimension=16 --nb_run=5
python train.py --dataset=citeseer --features=True --model=deep_gcn_vae --task=link_prediction --epochs=200 --learning_rate=0.01 --hidden=32 --dimension=16 --nb_run=5Pubmed
python train.py --dataset=pubmed --model=linear_ae --task=link_prediction --epochs=200 --learning_rate=0.01 --dimension=16 --nb_run=5
python train.py --dataset=pubmed --model=linear_vae --task=link_prediction --epochs=200 --learning_rate=0.01 --dimension=16 --nb_run=5
python train.py --dataset=pubmed --model=gcn_ae --task=link_prediction --epochs=200 --learning_rate=0.01 --hidden=32 --dimension=16 --nb_run=5
python train.py --dataset=pubmed --model=gcn_vae --task=link_prediction --epochs=200 --learning_rate=0.01 --hidden=32 --dimension=16 --nb_run=5
python train.py --dataset=pubmed --model=deep_gcn_ae --task=link_prediction --epochs=200 --learning_rate=0.01 --hidden=32 --dimension=16 --nb_run=5
python train.py --dataset=pubmed --model=deep_gcn_vae --task=link_prediction --epochs=200 --learning_rate=0.01 --hidden=32 --dimension=16 --nb_run=5Pubmed - with features
python train.py --dataset=pubmed --features=True --model=linear_ae --task=link_prediction --epochs=200 --learning_rate=0.01 --dimension=16 --nb_run=5
python train.py --dataset=pubmed --features=True --model=linear_vae --task=link_prediction --epochs=200 --learning_rate=0.01 --dimension=16 --nb_run=5
python train.py --dataset=pubmed --features=True --model=gcn_ae --task=link_prediction --epochs=200 --learning_rate=0.01 --hidden=32 --dimension=16 --nb_run=5
python train.py --dataset=pubmed --features=True --model=gcn_vae --task=link_prediction --epochs=200 --learning_rate=0.01 --hidden=32 --dimension=16 --nb_run=5
python train.py --dataset=pubmed --features=True --model=deep_gcn_ae --task=link_prediction --epochs=200 --learning_rate=0.01 --hidden=32 --dimension=16 --nb_run=5
python train.py --dataset=pubmed --features=True --model=deep_gcn_vae --task=link_prediction --epochs=200 --learning_rate=0.01 --hidden=32 --dimension=16 --nb_run=5Notes:
- Set
--task=node_clusteringwith same hyperparameters to evaluate models on node clustering (as in Table 4) instead of link prediction - Set
--nb_run=100to report mean AUC and AP along with standard errors over 100 runs, as in the paper - We recommend GPU usage for faster learning
Please cite our paper(s) if you use this code in your own work:
@misc{salha2019keep,
title={Keep It Simple: Graph Autoencoders Without Graph Convolutional Networks},
author={Salha, Guillaume and Hennequin, Romain and Vazirgiannis, Michalis},
howpublished={Workshop on Graph Representation Learning, 33rd Conference on Neural Information Processing Systems (NeurIPS)},
year={2019}
}@inproceedings{salha2019degeneracy,
title={A Degeneracy Framework for Scalable Graph Autoencoders},
author={Salha, Guillaume and Hennequin, Romain and Tran, Viet Anh and Vazirgiannis, Michalis},
booktitle={28th International Joint Conference on Artificial Intelligence (IJCAI)},
year={2019}
}