HAPNEST enables you to
- Efficiently generate large-scale, diverse and realistic datasets for genotypes and phenotypes
- Easily analyse data quality with an extensive workflow for evaluating synthetic data reliability and generalisability
- Examine the posterior distributions of model parameters to aid with model selection
We have used HAPNEST to simulate genetics data and corresponding phenotypes for 1 million individuals for 6 superpopulations and over 6.8 million variants in less than 12 hours (32GB RAM, 8 threads per machine). We have made this synthetic dataset (and a smaller example dataset) publicly available for download at https://www.ebi.ac.uk/biostudies/studies/S-BSST936.
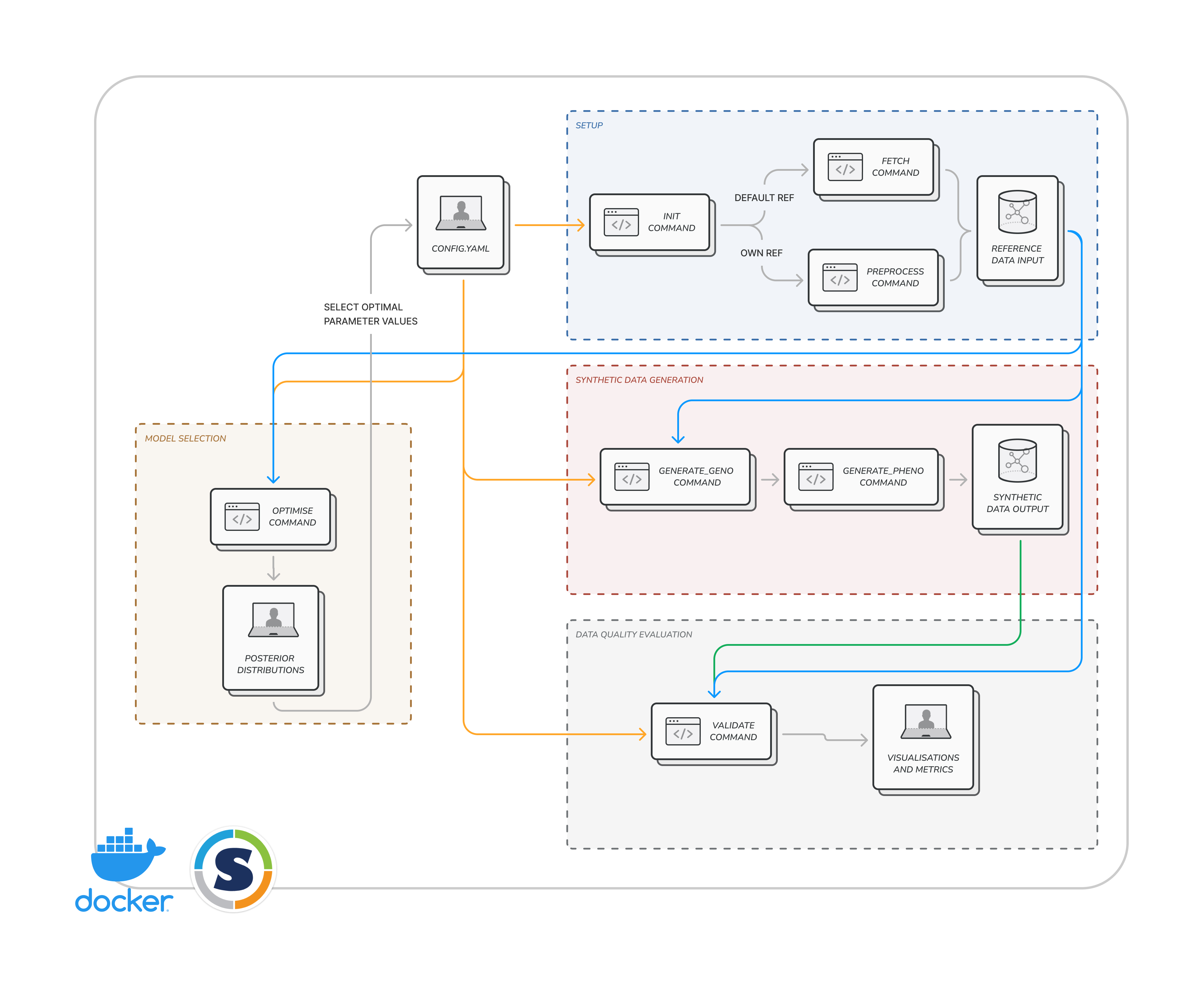
HAPNEST has a modular architecture design, where each software module can be used independently with a single command line call. This means, for example, you can use the evaluation workflow for evaluating synthetic data quality for any synthetic dataset, not just those generated by HAPNEST.
This software tool was developed by members of INTERVENE (INTERnational consortium of integratiVE geNomics prEdiction). If you would like to cite this software, please cite our preprint https://www.biorxiv.org/content/10.1101/2022.12.22.521552v1.
- Quickstart tutorial
- Customising your synthetic datasets
- Understanding your synthetic datasets
- Large scale synthetic data generation
Steps 1-4 only need to be run once the first time you install HAPNEST. Step 5 describes how you can generate and then evaluate a synthetic dataset. If you encounter any errors when running these steps, please see the more detailed documentation below.
- Download the Singularity container by running the following command:
singularity pull docker://sophiewharrie/intervene-synthetic-data
- Setup the directory structure illustrated below with the container you just downloaded and a copy of the
config.yamlfile from this repository:
.
├── data
| └── config.yaml
└── containers
└── intervene-synthetic-data_latest.sif
- Initialise the software dependencies from the root directory you set up in the previous step:
singularity exec --bind data/:/data/ containers/intervene-synthetic-data_latest.sif init
- Fetch the reference dataset:
singularity exec --bind data/:/data/ containers/intervene-synthetic-data_latest.sif fetch
- Generate synthetic genotypes and phenotypes, and evaluate the data quality:
singularity exec --bind data/:/data/ containers/intervene-synthetic-data_latest.sif generate_geno 1 data/config.yaml
singularity exec --bind data/:/data/ containers/intervene-synthetic-data_latest.sif generate_pheno data/config.yaml
singularity exec --bind data/:/data/ containers/intervene-synthetic-data_latest.sif validate data/config.yaml
This quickstart tutorial will show you the simplest approach for generating and evaluating synthetic datasets. This example will generate a small dataset of 1000 samples for 6 ancestry groups and HapMap3 variants on chromosome 1. Later we discuss how to customise the synthetic datasets.
- Download the HAPNEST container.
For ease of portability and reproducibility, we've made this software available as Docker and Singularity containers. Containerisation streamlines software dependency management by creating a standardised environment, to make it easier for you to get started with generating synthetic datasets.
The HAPNEST container is available at the link https://hub.docker.com/r/sophiewharrie/intervene-synthetic-data. The instructions provided in this documentation use the Singularity container, but can be adapted to work directly with the Docker container.
Download the Singularity container by running the following command:
singularity pull docker://sophiewharrie/intervene-synthetic-data
Alternatively, you can run this software without a container by manually installing the software dependencies. If you prefer this approach, you can view the list of required dependencies in the Dockerfile of this repository.
- Choose a location where you want to generate synthetic data, and create a
datadirectory containing a copy of theconfig.yamlfile downloaded from this repository and acontainersdirectory containing the downloaded container file.
.
├── data
| └── config.yaml
└── containers
└── intervene-synthetic-data_latest.sif
If your container version has a different name you should update this in any subsequent commands. The config.yaml file contains the parameter values used for synthetic data generation. In this tutorial we will use the default configuration.
- From the root directory you setup in the previous step, run the
initcommand to complete the setup of software dependencies:
singularity exec --bind data/:/data/ containers/intervene-synthetic-data_latest.sif init
This command binds the data directory you created in the previous step, which is used by HAPNEST to access input reference files and store output data files.
By default the above command will store Julia package files in the ~/.julia directory. If you experience an error related to updating the registry at ~/.julia/... you may need to give an explicit depot path by adding --env JULIA_DEPOT_PATH={path-to-your-preffered-location} to the previous command and all subsequent commands. For example:
singularity exec --bind data/:/data/ --env JULIA_DEPOT_PATH=/data/.julia containers/intervene-synthetic-data_latest.sif init
- The first time using the container, you need to fetch the reference dataset using the
fetchcommand:
singularity exec --bind data/:/data/ containers/intervene-synthetic-data_latest.sif fetch
This will download the reference files used as input to the synthetic data generation program to the data/inputs/processed directory. See preprocessing/README.md if you would like to know more about how this data was created and for information about creating your own reference datasets.
- Now generate a synthetic dataset using the
generate_genoandgenerate_phenocommands:
singularity exec --bind data/:/data/ containers/intervene-synthetic-data_latest.sif generate_geno 1 data/config.yaml
singularity exec --bind data/:/data/ containers/intervene-synthetic-data_latest.sif generate_pheno data/config.yaml
The number given after generate_geno in the first command is the number of computing threads you want the software to use. We recommend increasing this to a higher value for faster synthetic data generation. Running the above commands should generate a small synthetic dataset in the data/outputs/test directory.
Now it would be useful to evaluate the quality of this data.
- Evaluate synthetic data quality using the
validatecommand:
singularity exec --bind data/:/data/ containers/intervene-synthetic-data_latest.sif validate data/config.yaml
This will store visualisations in the data/outputs/test/evaluation directory and print results for quantitative metrics. See our manuscript for details about the evaluation workflow.
Now that you understand the basics, you can read about how to customise your synthetic datasets. You may also be interested in how to generate very large datasets.
You can customise the specifications for synthetic data generation by modifying the config.yaml file.
The global parameters are used by all workflows:
| Parameter name | Possible values | Description |
|---|---|---|
random_seed |
Integer value, e.g. 123 | A random seed used for reproducibility. |
chromosome |
all or an integer value between 1 and 22 |
Set to a numerical value between 1 and 22 if running the pipeline for a specific chromosome, or set to all for all 22 chromosomes. The all option will run synthetic data generation for each chromosome sequentially, so we recommend using the approach described in the large scale synthetic data generation section for efficiently generating multi-chromosome datasets. |
superpopulation |
none or one of AFR (African), AMR (Admixed American), EAS (East Asian), EUR (European), CSA (Central or South Asian), MID (Middle Eastern) |
A superpopulation for (all) synthetic samples. This parameter is ignored if using a custom population structure. |
memory |
Integer value | Amount of memory available (in MB) for memory-intensive commands. |
batchsize |
Integer value | The number of synthetic samples to write per batch, used during synthetic genotype generation. |
The most important filepaths to check before generating synthetic datasets are the general filepaths. Most other filepaths can be left with the default values.
The general filepaths are used by all workflows:
| Parameter name | Possible values | Description |
|---|---|---|
output_dir |
String | Directory for synthetic data generation outputs, e.g. data/outputs/test |
output_prefix |
String | Prefix for synthetic data generation outputs, using the {chromosome} wildcard, e.g. test_chr-{chromosome}. Please note that the program expects input files with the naming convention {prefix}-{chromosome} |
Filepaths for genotype data generation. You do not need to change these if you are using the reference data downloaded by the fetch command.
| Parameter name | Possible values | Description |
|---|---|---|
vcf_input_raw |
String | VCF files for the (real) reference dataset, before pre-processing |
vcf_input_processed |
String | VCF files for the (real) reference dataset, created by pre-processing |
vcf_metadata |
String | Text file describing metadata for the VCF reference such as name of SNPs |
popfile_raw |
String | Population file for the reference VCF, before pre-processing |
popfile_processed |
String | Population file for the reference VCF, created by pre-processing |
variant_list |
String | List of variants to include in synthetic dataset |
remove_list |
String | List of samples to remove from the reference dataset |
rsid_list |
String | Map of variant names to rsID format |
genetic_mapfile |
String | Genetic maps for converting basepair to centimorgan distances |
genetic_distfile |
String | A genetic map created by the pre-processing code |
mutation_mapfile |
String | Age of mutation for each variant |
mutation_agefile |
String | A mutation age map created by the pre-processing code |
hap1_matrix |
String | A data structure for the reference data haplotypes created by the pre-processing code |
hap2_matrix |
String | A data structure for the reference data haplotypes created by the pre-processing code |
Filepaths for phenotype data generation. See the phenotype data parameters section for other parameters that can be specified to customise the phenotype generation.
| Parameter name | Possible values | Description |
|---|---|---|
causal_list |
String | Filepath for a list of predefined SNPs to be used as causal, overrides polygenicity parameter if specified. Each column contains causal SNPs for one trait, columns separated by comma. |
reference_list |
String | Filepath for a reference file for LD. By default, uses the reference file downloaded by the fetch command. |
plink_override |
String | Optional argument which can be used if you would like to generate phenotypes for a genetics dataset that is in a different location to what is given by output_dir and output_prefix. Specify the location of the PLINK files with the naming convention {prefix}-{chromosome} |
Do not change these if you are using the Docker/Singularity containers. However, if you manually installed the software dependencies then you should check and update these filepaths:
| Parameter name | Possible values | Description |
|---|---|---|
plink |
String | Software path for plink software |
plink2 |
String | Software path for plink2 software |
king |
String | Software path for king software |
vcftools |
String | Software path for vcftools software |
mapthin |
String | Software path for mapthin software |
phenoalg |
String | Software path for our own phenotype software |
There are three approaches for generating synthetic genotype data:
| Approach | Result | Instructions |
|---|---|---|
| Same superpopulation for all samples | Generates synthetic samples for a specific superpopulation group | 1. Set global_parameters > superpopulation to your selected superpopulation, 2. Set genotype_data > samples > default to true, 3. Set genotype_data > default > nsamples to the number of samples you want to generate |
| Equal ratio of samples for all six superpopulations | Generates synthetic samples for all six superpopulation groups in equal ratio, e.g. 1000 samples for each superpopulation if nsamples is 6000 |
1. Set global_parameters > superpopulation to none, 2. Set genotype_data > samples > default to true, 3. Set genotype_data > default > nsamples to the number of samples you want to generate |
| Custom population structure | Generates synthetic samples according to your own custom population structure | 1. Set genotype_data > samples > default to false (this will cause the algorithm will ignore the value of the global_parameters > superpopulation parameter), 2. See the section on configuring custom population structure for how to define custom population structure |
The genotype algorithm also uses a recombination rate rho and effective population size Ne. The default values for each superpopulation were selected as the means of the posterior distributions derived using the optimisation workflow, as described in our manuscript. You can override these values in the configuration file.
Customise the population structure of the synthetic dataset by specifying your own population groups. You can add as many population groups as you want.
A population group consists of:
id: a name for your population group, e.g.pop1nsamples: the number of samples you want to generate for this population group, e.g. 100populations: the population structure for the population group, as a list of superpopulations that you want to include, and the proportion of segments from each superpopulation for each synthetic genotype (which must sum to 100)
Example 1: 100 genotypes with EUR ancestry and 200 genotypes with AFR ancestry
- id: EUR_pop
nsamples: 100
populations:
- EUR: 100
- id: AFR_pop
nsamples: 200
populations:
- AFR: 100
Example 2: 100 genotypes where each genotype has 50% of segments sampled from EUR reference samples and 50% of segments sampled from AFR reference samples. Please note that this does not result in true admixed genotypes because the current implementation of the algorithm does not account for the time of admixture events.
- id: admix_pop
nsamples: 100
populations:
- EUR: 50
- AFR: 50
Parameters for phenotype data generation:
| Parameter name | Possible values | Description |
|---|---|---|
| nPopulation | Integer | The number of populations (nPop) |
| nTrait | Integer | The number of traits |
| a | Float | Suggested value is -0.4 |
| b | Float | Suggested value is -1 |
| c | Float | Suggested value is 0.5 |
| nComponent | Integer | Number of Gaussian mixture components |
| ProportionGeno | Comma-separated list of floats | The observed causal SNP heritability in each population, each trait. Flatten nPop * nTrait matrix, entries separated by comma |
| ProportionCovar | Comma-separated list of floats | The observed proportion of variance contributed by the covariate (input in SampleList file) in each population, each trait. Flatten nPop * nTrait matrix, entries separated by comma. |
| Prevalence | Comma-separated list of floats | Disease prevalence in each population, each trait. Flatten nPop * nTrait matrix, entries separated by comma. If prevalence is specified, output will include a column for binary case/control statues. |
| TraitCorr | Comma-separated list of floats | A flattened correlation matrix for traits genetic correlation (symmetric positive definite). nTrait x nTrait entries separated by comma. |
| PopulationCorr | Comma-separated list of floats | A flattened correlation matrix for population genetic correlation (symmetric positive definite). nPop x nPop entries separated by comma. |
| CompWeight | Comma-separated list of floats | Gaussian mixture component weights |
| UseCausalList | Boolean | True if using a list of predefined SNPs to be used as causal, overrides Polygenicity parameter if specified. Each column contains causal SNPs for one trait, columns separated by comma. See phenotype filepaths for how to specify a causal list as input. |
| Polygenicity | Float | nTrait vector of trait polygenicity, measured by proportion of total SNPs being causal. e.g. Polygenicity = 0.05 meaning around 5% SNPs will be causal. |
| Pleiotropy | Float | nTrait vector of trait's pleiotropy relationship comparing to trait 1. i.e. if trait 2 has Pleiotropy = 0.9, it means 90% of causal SNPs in trait 1 are also causal in trait 2. Therefore, first entry of Pleiotropy vector is always 1. Entries separated by comma. |
The evaluation workflow computes a set of visualisations and quantitative metrics for understanding the reliability and generalisability of a synthetic dataset. This is done by a chromosome-by-chromosome comparison of the synthetic data with a real reference dataset from multiple perspectives (LD structure, kinship relatedness structure, minor allele frequencies, etc.).
The evaluation workflow can be run using the validate command, e.g.
singularity exec --bind data/:/data/ containers/intervene-synthetic-data_latest.sif validate data/config.yaml
Note that you can use the evaluation workflow with any PLINK-formatted synthetic dataset, i.e. it doesn't have to be generated using this tool.
You can enable/disable different types of metrics by setting true/false values in the configuration file:
| Parameter name | Possible values | Description | Requires genotype data | Requires phenotype data |
|---|---|---|---|---|
aats |
true/false |
Nearest neighbour adversarial accuracy, which quantifies the extent to which synthetic data can be statistically distinguished from real data | ✔️ | |
kinship |
true/false |
Kinship-based relatedness analysis, including kinship density and identical by segment (IBS) plots | ✔️ | |
ld_corr |
true/false |
Linkage disequilibrium (LD) correlation matrix | ✔️ | |
ld_decay |
true/false |
Linkage disequilibrium (LD) decay plot (and distance) | ✔️ | |
maf |
true/false |
Minor allele frequency divergences | ✔️ | |
pca |
true/false |
Principal components analysis of population structure | ✔️ | |
gwas |
true/false |
Run GWAS and generate manhattan and qqplot | ✔️ | ✔️ |
The evaluation metrics are printed to standard output and plots are stored at output_dir/evaluation, where output_dir is specified in the config.yaml file.
The optimisation workflow uses approximate Bayesian computation (ABC) rejection sampling to estimate the posterior distributions of the unknown model parameters for the genotype generation model. This workflow can be run using the optimise command, e.g.
singularity exec --bind data/:/data/ containers/intervene-synthetic-data_latest.sif optimise data/config.yaml
| Parameter name | Possible values | Description |
|---|---|---|
priors |
Numerical values for uniform_lower and uniform_upper of each parameter |
The lower and upper bounds for the uniform prior distributions of each parameter. |
simulation_rejection_ABC |
Set run to true if using this algorithm. Specify the number of posterior samples with n_particles, the maximum number of simulations to run with max_iter, the acceptance threshold with threshold, and whether the algorithm progress should be printed to standard output with write_progress. |
Configuration for simulation-based rejection sampling. |
emulation_rejection_ABC |
Set run to true if using this algorithm. Specify the number of posterior samples with n_particles, the number of samples for training the emulator with n_design_points, the maximum number of emulations to run with max_iter, the acceptance threshold with threshold, and whether the algorithm progress should be printed to standard output with write_progress. |
Configuration for emulation-based rejection sampling. Recommended for computationally expensive simulations of large synthetic datasets. |
summary_statistics |
Set true/false for ld_decay and/or kinship |
The ld_decay objective minimises Euclidean distance between LD decay curves of the reference/synthetic datasets. The kinship objective minimises genetic relatedness between the reference/synthetic datasets. The objectives can be used separately or combined together. |
In this section we provide instructions for utilising a HPC cluster to efficiently generate very large datasets. These instructions assume that your cluster uses the Slurm job scheduler.
- Create a
config.yamlfile with the parameters you want to use for synthetic data generation. Specify thechromosomeparameter as a wildcard as shown below to enable the software to efficiently generate data for all 22 chromosomes simultaneously.
global_parameters:
chromosome: ${chr}
...
- The script below shows an example of a batch script that can be used to submit a collection of jobs for generating synthetic genotypes. This script will copy the configuration file for each chromosome and submit 22 multi-threaded jobs. You can adjust the computational resources for your use case - the example below shows what we used to generate 1 million genotypes for over 6.8 million variants.
#!/bin/bash
#SBATCH --array=1-22
#SBATCH --mem-per-cpu=32G
#SBATCH --cpus-per-task=8
#SBATCH --time 24:00:00
n=$SLURM_ARRAY_TASK_ID
CONFIG=data/config # prefix for config file
# generate a config for each chromosome
cp ${CONFIG}.yaml ${CONFIG}$n.yaml
sed -i 's/${chr}'"/$n/g" ${CONFIG}$n.yaml
# generate data for each chromosome
singularity exec --bind data/:/data/ containers/intervene-synthetic-data_latest.sif generate_geno 8 ${CONFIG}$n.yaml
- The script below shows how to generate synthetic phenotypes, using the genotypes generated in the previous step as input. This shouldn't be split into multiple jobs because the phenotype generation algorithm needs to sum genetic effects across all chromosomes included in the dataset.
#!/bin/bash
CONFIG=data/config # prefix for config file
# replace the chromosome wildcard with all
cp ${CONFIG}.yaml ${CONFIG}_pheno$n.yaml
sed -i 's/${chr}'"/all/g" ${CONFIG}_pheno$n.yaml
# generate phenotype data
singularity exec --bind data/:/data/ containers/intervene-synthetic-data_latest.sif generate_pheno ${CONFIG}_pheno$n.yaml