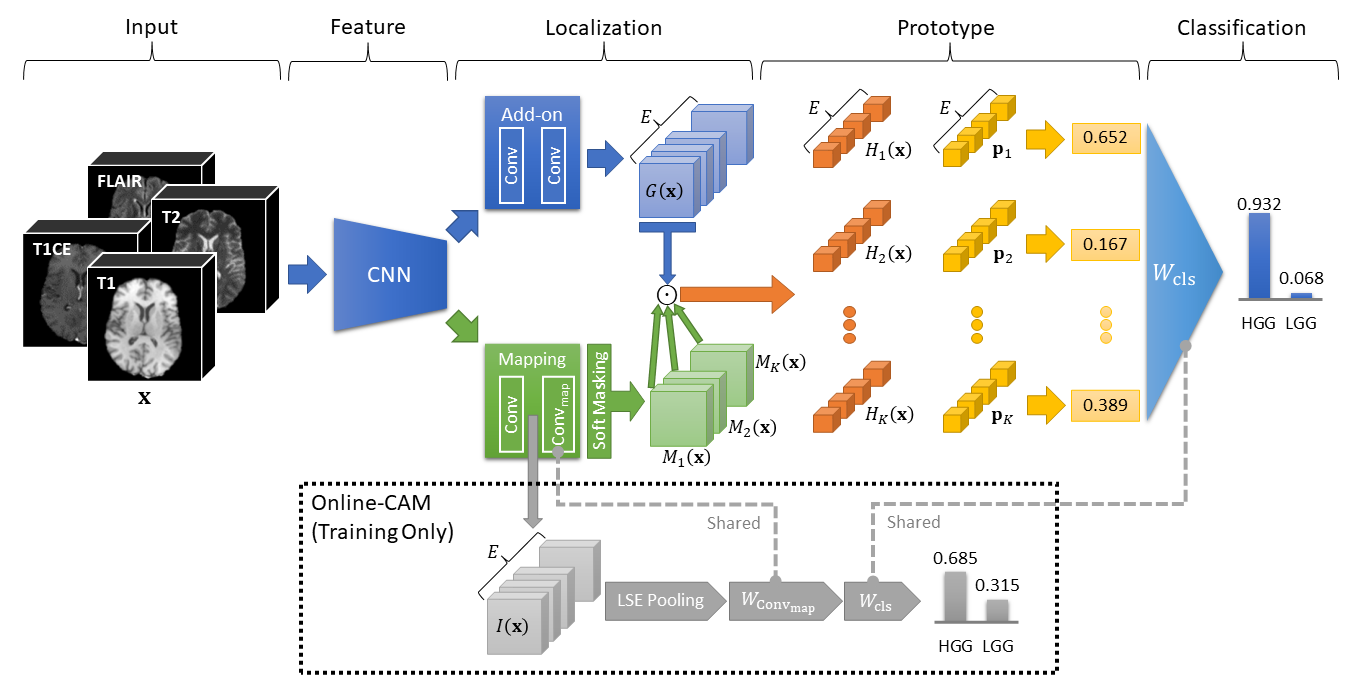
This repository contains the official implementation of MProtoNet from the paper "MProtoNet: A Case-Based Interpretable Model for Brain Tumor Classification with 3D Multi-parametric Magnetic Resonance Imaging" (accepted at MIDL 2023) by Yuanyuan Wei, Roger Tam, and Xiaoying Tang.
Talk: https://www.youtube.com/watch?v=DtfXwrliVQg
- Directory Structure
- Dataset
- Environment Configuration
- Experiments
- Results
- Acknowledgment
- Citation
- To-do List
data/: Datasetenv_config/: Environment configurationresults/: Resultssrc/: Source codetumor_cls.py: Major code for running the experiments
Please follow the instructions on the website above to request the BraTS_2020 dataset (it may take 1~4 days) and download MICCAI_BraTS2020_TrainingData.zip to the data/ folder. Then unzip the dataset:
unzip ./data/MICCAI_BraTS2020_TrainingData.zip -d ./data/BraTS_2020Prerequisites: Python (>=3.8), NumPy, SciPy, scikit-learn, Matplotlib, Jupyter, PyTorch, Captum, OpenCV, TorchIO.
If you are using Miniconda/Anaconda, you can run following commands (Linux & Windows) to create a new conda environment called mprotonet with all the required packages (they can take up about 10 GB of disk space).
conda env create -f ./env_config/environment.yml -n mprotonet -y
# Or if you want to install the latest versions of the packages (with Python 3.11 and CUDA 11):
# conda env create -f ./env_config/environment_latest.yml -n mprotonet -y
conda activate mprotonet
conda clean -a -y
conda env listThe following experiments can consume about 12~18 GB of GPU memory.
cd ./src
# CNN (with GradCAM)
python ./tumor_cls.py -m CNN3D -n 100 -p "{'batch_size': [32], 'lr': [0.001], 'wd': [0.01], 'features': ['resnet152_ri'], 'n_layers': [6]}" --save-model 1
# ProtoPNet
python ./tumor_cls.py -m MProtoNet3D_pm1 -n 100 -p "{'batch_size': [32], 'lr': [0.001], 'wd': [0.01], 'features': ['resnet152_ri'], 'n_layers': [6], 'prototype_shape': [(30, 128, 1, 1, 1)], 'f_dist': ['cos'], 'topk_p': [1], 'coefs': [{'cls': 1, 'clst': 0.8, 'sep': -0.08, 'L1': 0.01, 'map': 0, 'OC': 0}]}" --save-model 1
# XProtoNet
python ./tumor_cls.py -m MProtoNet3D_pm2 -n 100 -p "{'batch_size': [32], 'lr': [0.001], 'wd': [0.01], 'features': ['resnet152_ri'], 'n_layers': [6], 'prototype_shape': [(30, 128, 1, 1, 1)], 'f_dist': ['cos'], 'topk_p': [1], 'coefs': [{'cls': 1, 'clst': 0.8, 'sep': -0.08, 'L1': 0.01, 'map': 0.5, 'OC': 0}]}" --save-model 1
# MProtoNet A
python ./tumor_cls.py -m MProtoNet3D_pm3 -n 100 -p "{'batch_size': [32], 'lr': [0.001], 'wd': [0.01], 'features': ['resnet152_ri'], 'n_layers': [6], 'prototype_shape': [(30, 128, 1, 1, 1)], 'f_dist': ['cos'], 'topk_p': [1], 'coefs': [{'cls': 1, 'clst': 0.8, 'sep': -0.08, 'L1': 0.01, 'map': 0.5, 'OC': 0}]}" --save-model 1
# MProtoNet B
python ./tumor_cls.py -m MProtoNet3D_pm4 -n 100 -p "{'batch_size': [32], 'lr': [0.001], 'wd': [0.01], 'features': ['resnet152_ri'], 'n_layers': [6], 'prototype_shape': [(30, 128, 1, 1, 1)], 'f_dist': ['cos'], 'topk_p': [1], 'coefs': [{'cls': 1, 'clst': 0.8, 'sep': -0.08, 'L1': 0.01, 'map': 0.5, 'OC': 0.05}]}" --save-model 1
# MProtoNet C
python ./tumor_cls.py -m MProtoNet3D_pm5 -n 100 -p "{'batch_size': [32], 'lr': [0.001], 'wd': [0.01], 'features': ['resnet152_ri'], 'n_layers': [6], 'prototype_shape': [(30, 128, 1, 1, 1)], 'f_dist': ['cos'], 'topk_p': [1], 'coefs': [{'cls': 1, 'clst': 0.8, 'sep': -0.08, 'L1': 0.01, 'map': 0.5, 'OC': 0.05}]}" --save-model 1This repository contains modified source code from cfchen-duke/ProtoPNet (MIT License) by Chaofan Chen, Oscar Li, Chaofan Tao, Alina Jade Barnett, and Cynthia Rudin.
Yuanyuan Wei, Roger Tam, and Xiaoying Tang. MProtoNet: a case-based interpretable model for brain tumor classification with 3D multi-parametric magnetic resonance imaging. In Medical Imaging with Deep Learning, Nashville, United States, July 2023.
@inproceedings{wei2023mprotonet,
title = {{{MProtoNet}}: A Case-Based Interpretable Model for Brain Tumor Classification with {{3D}} Multi-Parametric Magnetic Resonance Imaging},
shorttitle = {{{MProtoNet}}},
booktitle = {Medical {{Imaging}} with {{Deep Learning}}},
author = {Wei, Yuanyuan and Tam, Roger and Tang, Xiaoying},
year = {2023},
month = jul,
address = {{Nashville, United States}},
url = {https://openreview.net/forum?id=6Wbj3QCo4U4}
}- Dataset
- Environment configuration
- Upload source code
- Experiments
- Results