RetinaFed: Exploring Novel Techniques for Predicting Diabetic Retinopathy through Federated Learning
In this project, we aim to explore different federated learning approaches to train distributed models on private healthcare data. We explore the federated learning setup for diabetic retinopathy area and delve deep into how different data distributions can affect the performance of our models
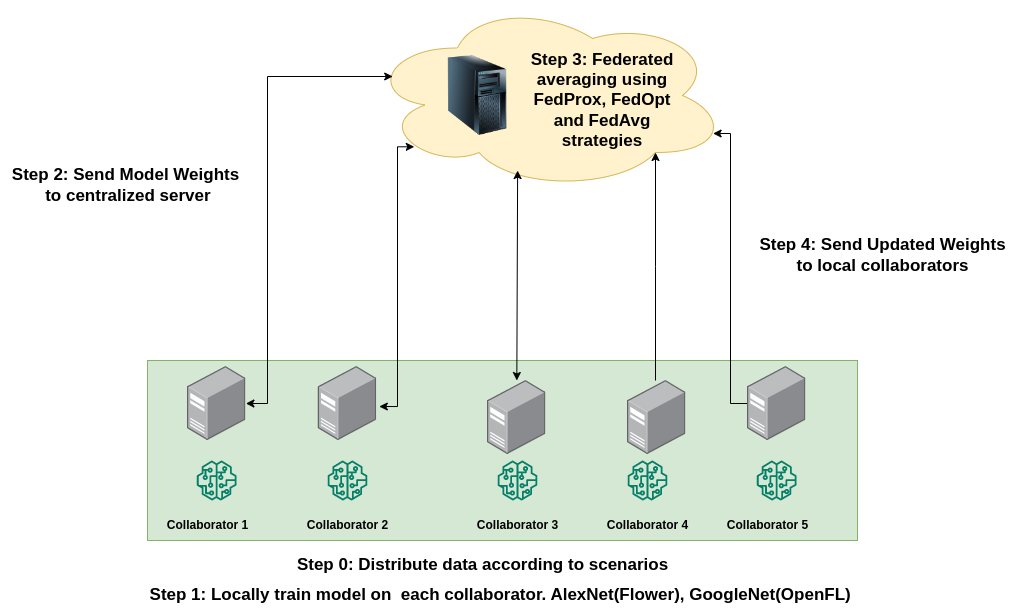
The proposed Federated Learning process consists of 3 steps :-
- Local Training - Each Hospital serves as a local client that trains its local models on a unique dataset it hosts. The weights of the trained model are shared with a central server.
- Global Aggregration - The centralized server is connected with several clients (hospitals) and receives a locally trained model from each of them. To ensure further privacy, these weights can be encrypted while being sent over the network. The central server uses several strategies to create a globalized model from all the local models.
- Evaluation - The globalized model is evaluated on a validation/test set to determine F1 score. Further, it is sent to each of the hospitals/clients and replaces the local model.
The globalized model can be created using several strategies. We explore FedAvg, FedProx and FedOpt strategies in our work.
- Dataset: Diabetic Retinopathy 224x224 Gaussian Filtered
- Source: Kaggle
- Training-Validation Split: 80%-20%
- Length of Dataset: 3662 samples of (224 * 224)
- Classes:
- No Diabetic Retinopathy: 1805
- Mild: 370
- Moderate: 999
- Severe: 193
- Proliferative Diabetic Retinopathy: 295
The table illustrates the different scenarios of data distribution that we experimented on
| Nomenclature | Cases | Classes per Client | Length per Client |
|---|---|---|---|
| Case 1 | Easiest Case (I.I.D) | Equal (=5) | Equal |
| Case 2 | Easy Case | Random | Equal |
| Case 3 | Average Case | Random | Random |
| Case 4 | Average Case | 4 | Random |
| Case 5 | Average Case | 3 | Random |
| Case 6 | Hard Case | 2 | Random |
| Case 7 | Hardest Case | 1 | Random |
| OpenFL | Flower | |
|---|---|---|
| Model | GoogleNet (Inception-V1) with single FC layer | AlexNet with single FC layer |
| Learning Rate | 0.001 | 0.001 |
| Optimizer | Adam (FedOpt, FedAvg) | Adam (FedOpt, FedProx, FedAvg) |
| Mu | 0.8 (FedProx) | 0.8 (FedProx) |
| Number of Collaborators / Clients | 5 | 5 |
| Implementation Library | PyTorch | PyTorch |
| Learnable Parameters | 5125 | 20485 |
| Batch Size | 16 | 8 |
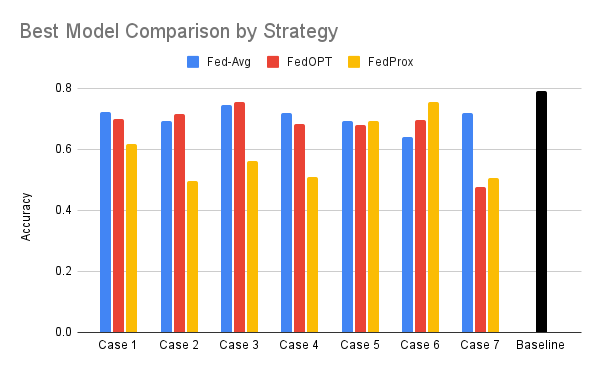
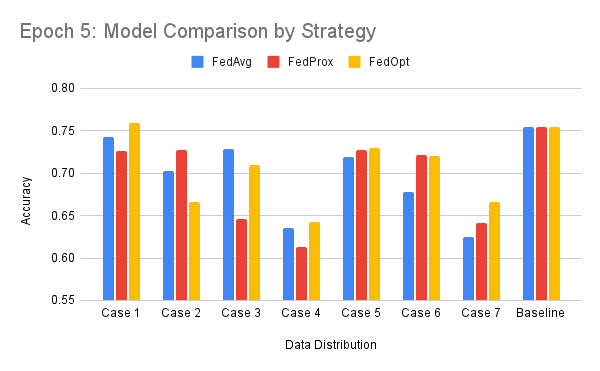
All values denote model accuracy on global validation set.
| FedAvg* | FedOPT* | FedProx* | |
|---|---|---|---|
| Case 1 | 0.722372 | 0.699015 | 0.61705 |
| Case 2 | 0.692098 | 0.71618 | 0.496689 |
| Case 3 | 0.75039 | 0.755319 | 0.747816 |
| Case 4 | 0.71875 | 0.683089 | 0.692641 |
| Case 5 | 0.692641 | 0.678082 | 0.720419 |
| Case 6 | 0.641992 | 0.696118 | 0.755319 |
| Case 7 | 0.72861 | 0.475783 | 0.507422 |
| Baseline | 0.791667 | ||
| FedAvg* | FedOPT* | FedProx* | |
|---|---|---|---|
| Case 1 | 0.722372 | 0.699015 | 0.61705 |
| Case 2 | 0.692098 | 0.71618 | 0.496689 |
| Case 3 | 0.75039 | 0.755319 | 0.747816 |
| Case 4 | 0.71875 | 0.683089 | 0.692641 |
| Case 5 | 0.692641 | 0.678082 | 0.720419 |
| Case 6 | 0.641992 | 0.696118 | 0.755319 |
| Case 7 | 0.72861 | 0.475783 | 0.507422 |
| Baseline | 0.791667 | ||
Download dataset from Kaggle: Link into the dataset folder. Perform Following operations in the main directory
cd dataset
unzip archive.zip
cd gaussian_filtered_images/gaussian_filtered_images/
All the OpenFL experiments can be running using OpenFL-Diabetic-Demo-CustomDataDistributor notebook In order to experiment on a particular data distribution use the data_splitter function.
import DataSplitterMethods
data_splitter = DataSplitterMethods.SplitFunctionGenerator("Equal-Equal-Split") #Use strings defined below to experiment on data distribution
# Pass this data splitter to fl_data in the train_splitter argument
fl_data = FederatedDataSet(train_data, train_labels, test_data, test_labels,
batch_size = batch_size , num_classes = num_classes,
train_splitter=data_splitter)In the notebook, you only need to change the string in the cell for data splitter methods and run the entire notebook
| Scenario | String |
| Case 1 | Equal-Equal-Split |
| Case 2 | Random-Equal-Split |
| Case 3 | Random-Unequal-Split |
| Case 4 | 4-Class-per-collab-split |
| Case 5 | 3-Class-per-collab-split |
| Case 6 | 2-Class-per-collab-split |
| Case 7 | 1-Class-per-collab-split |
Note: Google Colab and Google Drive are needed for executing the Diabetic Retinopathy experiments using the Flower federated learning framework. This environment choice is dictated by the memory requirements and installation dependencies.
Download dataset from Kaggle: Link into an archive folder. Upload the archive folder to your Google Drive. The resulting file structure will be as follows:
All data distribution experiments for Flower FedAvg aggregation strategy can be executed by running the /Flower/Flower_Diabetic_Demo_Custom_Model_Fed_Avg_With_Images.ipynb notebook.
All data distribution experiments for Flower FedProx aggregation strategy can be executed by running the /Flower/Flower_Diabetic_Demo_Custom_Model_Fed_Prox_With_Images.ipynb notebook.
All data distribution experiments for Flower FedOpt aggregation strategy can be executed by running the /Flower/Flower_Diabetic_Demo_Custom_Model_Fed_Opt_With_Images.ipynb notebook.