Overview
Installation
Tutorial
User Guide
Recommended workflow
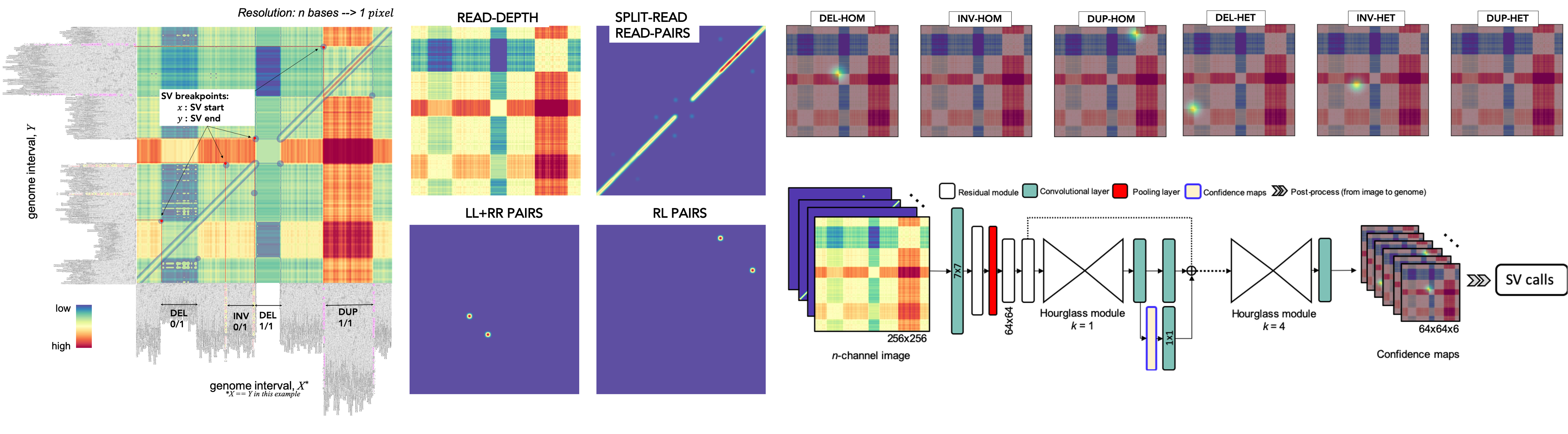
Cue is a deep learning framework for SV calling and genotyping. At a high-level, Cue operates in the following stages illustrated in the figure below:
- sequence alignments are converted into images that capture multiple alignment signals across two genome intervals,
- a trained neural network is used to generate Gaussian response confidence maps for each image, which encode the location, type, and genotype of the SVs in this image, and
- the high-confidence SV predictions are refined and mapped back from image to genome coordinates.
The current version of Cue can be used to detect and genotype the following SV types: deletions (DELs), tandem duplication (DUPs), inversions (INVs), deletion-flanked inversions (INVDELs), and inverted duplications (INVDUPs) larger than 5kbp.
For more information please see the following preprint and video.
- Clone the repository:
git clone git@github.com:PopicLab/cue.git
-
Create the virtual environment (in the env directory):
$> python3.7 -m venv env -
Activate the environment:
$> source env/bin/activate -
Install all the required packages in the virtual environment (this should take a few minutes):
$> pip --no-cache-dir install -r install/requirements.txt
Packages can also be installed individually using the recommended versions provided in theinstall/requirements.txtfile; for example:$> pip install numpy==1.18.5 -
Set the
PYTHONPATHas follows:export PYTHONPATH=${PYTHONPATH}:/path/to/cue
To deactivate the environment: $> deactivate
Alternatively, Cue can be installed using theDockerfile provided in the install/docker directory
from Cue's capsule in CodeOcean.
We recommend trying the provided demo Jupyter notebook to ensure that the software
was properly installed and to experiment running Cue. For convenience, Jupyter was already included in the installation
requirements above, or can be installed separately from here.
In this demo we use Cue to discover variants in a small BAM file provided here: data/demo/inputs/chr21.small.bam
(with the associated YAML config files needed to execute this workflow provided in the data/demo/config directory).
In addition to the functionality to call structural variants, the framework can be used to execute
custom model training, evaluation, and image generation. The engine directory contains the following
key high-level scripts to train/evaluate the model and generate image datasets:
call.py: calls structural variants given a pre-trained model and an input BAM file (can be executed on multiple GPUs or CPUs)train.py: trains a deep learning model (currently, this is a stacked hourglass network architecture) to detect SV keypoints in imagesgenerate.py: creates an annotated image dataset from alignments (BAM file(s))view.py: plots images annotated with SVs from a VCF/BED file given genome alignments (BAM format); can be used to visualize model predictions or ground truth SVs
Each script accepts as input one or multiple YAML config files, which encode a variety of parameters.
Template config files are provided in the config directory.
The key required and optional YAML parameters for each Cue command are listed below.
call.py (data YAML):
bam[required] path to the alignments BAM filefai[required] path to the referene FASTA FAI filen_cpus[optional] number of CPUs to use for calling (parallelized by chromosome)chr_names[optional] list of chromosomes to process: null (all) or a specific list e.g. ["chr1", "chr21"]
call.py (model YAML):
model_path[required] path to the pretrained Cue modelgpu_ids[optional] list of GPU ids to use for calling -- CPU(s) will be used if empty
train.py:
dataset_dirs[required] list of annotated imagesets to use for traininggpu_ids[optional] GPU id to use for training -- a CPU will be used if emptyreport_interval[optional] frequency (in number of batches) for reporting training stats and image predictions
generate.py:
bam[required] path to the alignments BAM filebed[required] path to the ground truth BED or VCF filefai[required] path to the referene FASTA FAI filen_cpus[optional] number of CPUs to use for image generation (parallelized by chromosome)chr_names[optional] list of chromosomes to process: null (all) or a specific list e.g. ["chr1", "chr21"]
view.py:
bam[required] path to the alignments BAM filebed[required] path to the BED or VCF file with SVs to visualizefai[required] path to the reference FASTA FAI filen_cpus[optional] number of CPUs (parallelized by chromosome)chr_names[optional] list of chromosomes to process: null (all) or a specific list e.g. ["chr1", "chr21"]
- Create a new directory.
- Place YAML config file(s) in this directory (see the provided templates).
- Populate the YAML config file(s) with the parameters specific to this experiment.
- Execute the appropriate
enginescript providing the path to the newly configured YAML file(s). The engine scripts will automatically create auxiliary directories with results in the folder where the config YAML files are located.
Victoria Popic (vpopic@broadinstitute.org)
For questions, suggestions, or technical assistance, please create an issue on the Cue Github issues page or reach out to Victoria Popic at vpopic@broadinstitute.org.