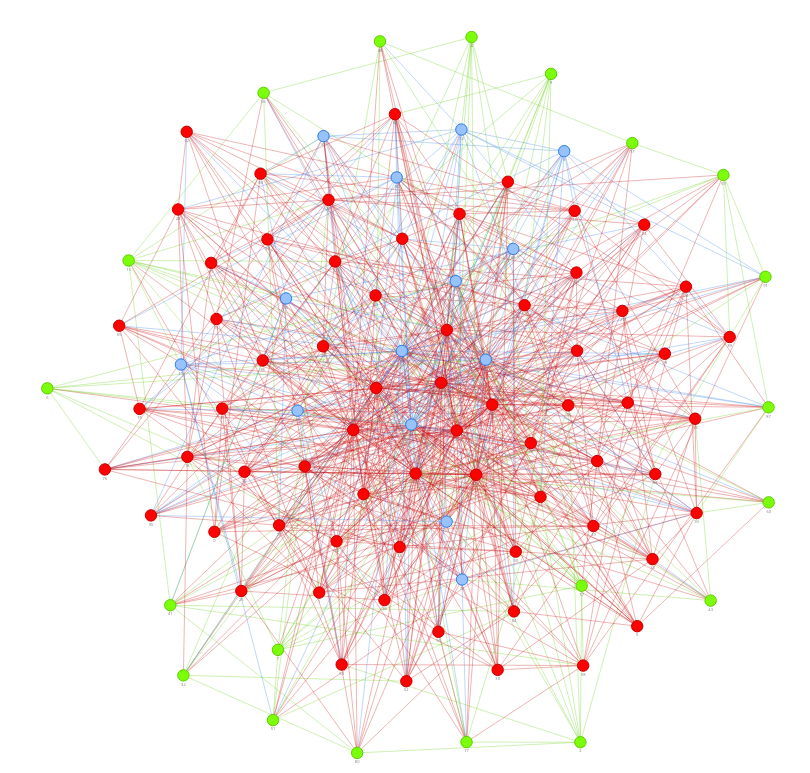
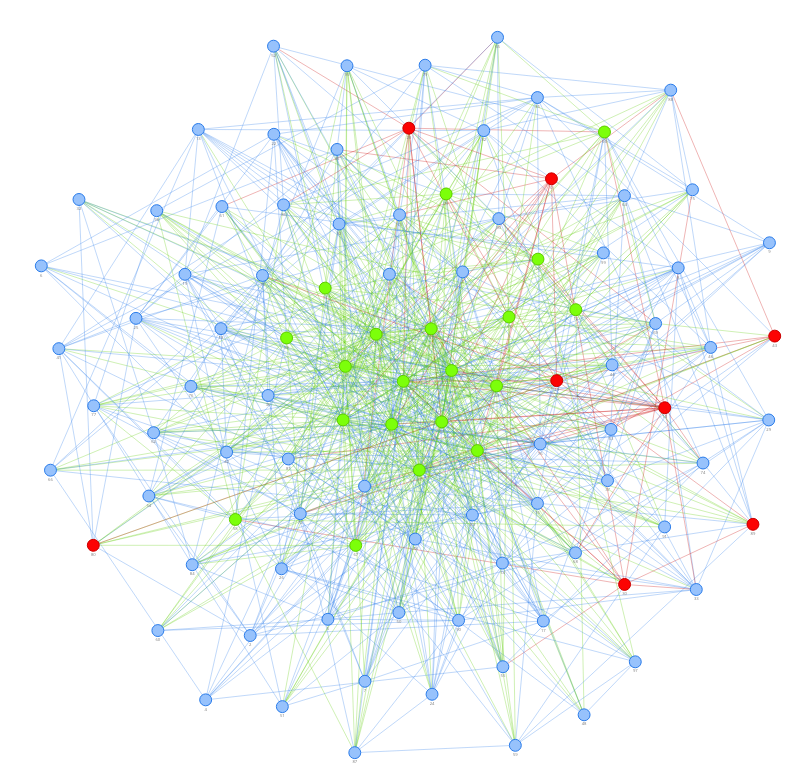
This is a simulator of COVID-19 transmission model constructed by pyvis.
We can visualize the result by adjusting the parameters.
- modified [SIR model] [1] (use vaccination instead of recovery).
- designed by multiprocessing pattern for computing efficiency improving on multiple CPUs machine.
- infectivity variate with time
- one person usually will interactive with the same people, and this model adopts this rule. However, we determine the contact people number by uniform probability (default between 0.9-1).
- the probability of be infected determinates by the patient's infectivity.
The 1st one illustrated the effect for vaccination on people who are not social active (for example, people quarantined at home), and the 2nd one is vaccination on those who contact others frequently (for example, convenience store clerks).
Obviously, the latter can protect more people who are not vaccinated.
The node color means:
- Red: infected
- Green: vaccinated
- Blue: neither infected nor vaccinated
The first seed is node 0.
Node in the middle area means more social activity with other persons and less in the marginal zone.
This model tells us that we might protect about 85% people by vaccinating 25% persons with [mRNA vaccines] [2] in the middle area if we take immediate action in the first 5-10 days.
If you have any idea for improving this project, please don’t hesitate to let me know.
# picture height
pxHeight='800px'
# width ratio relative to height
ratioWidth='100%'
# background color
colorbBg='#222222'
# node font color
colorNodeFont='white'
# how much processes will be used
iProcNum = 3
# how much chunks that total tasks will be divided
iChunksNum = iProcNum + 1
# wait seconds(for each proccess)
iWaitWkrSec = 86400
# color of infected nodes
hexInfColor = '#fc0303'
# color of vaccinated nodes
hexVaccColor = '#7cff0a'
# desired vaccinated ratio
floatVaccRatio = 0.2
# days without vaccination in the pandemic front stage
iHeadDaysNoVacc = 0
# days for vaccination activity in the pandemic
iVaccDays = 10
# days after vaccination activity
iTailDaysNoVacc = 0
# the 1st node be infected
iFirstInfPid = 0
# for random.uniform(low, up)
# random fine-tuning coefficient lower limit of maximum possible contact counts
floatInfRandFactorLow = 0.9
# random fine-tuning coefficient upper limit of maximum possible contact counts
floatInfRandFactorUpper = 1.0
# random selected max contacts number of one person (quarantine policy can be simulated here)
tupleMaxConn = (1, 3, 9, 49)
# weight for select one item of tupleMaxConn
tupleWtOfChoiceMaxConn = (2, 11, 13, 3)
# the dictionary used for infectivity (variated with days, for example, the infectivity on 4th day after infected is 40%)
# and you can simulate wearing face mask and effect of social distance here
dictInfectivity = {1: 0, 2: 0, 3: 0, 4: 0.6, 5: 0.7, 6: 0.6, 7: 0.4, 8: 0.2, 9: 0.1, 10: 0}
# dictInfectivity = {1: 0, 2: 0, 3: 0, 4: 0.1, 5: 0.1, 6: 0.1, 7: 0.1, 8: 0.1, 9: 0.1, 10: 0}
# Total population
iPopCnt = 500
- not consider mortality and its effect.
- Assuming that one won’t infect others immediately after a shot of vaccine. Recent data reveals that in an adjusted model, the viral RNA load was 40% lower (95% CI, 16.3 to 57.3) with at least partial vaccination than with no vaccination. [3] However, the relationship among viral load, viral shedding, and infectivity is not clear. If somebody get the data, please inform me.
- not for delta variant.
[1] https://www.maa.org/press/periodicals/loci/joma/the-sir-model-for-spread-of-disease-the-differential-equation-model "SIR model"
[2] https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/fully-vaccinated-people.html "mRNA vaccines"
[3] https://www.nejm.org/doi/full/10.1056/NEJMoa2107058?query=recirc_mostViewed_railB_article "viral RNA load"