
A pure-Python, PyTorch-like automatic differentiation library for education.
.
├── match # The Match library
│ ├── __init__.py # --- contains default import statements
│ ├── list2d.py # --- a storage class for matrix data
│ ├── matrix.py # --- a 2D matrix class (including autodiff)
│ └── nn.py # --- higher-level neural network functionality
├── demo_linear.ipynb # A linear model demo (Jupyter)
├── demo_linear.py # A linear model demo (script)
├── test.py # Unit-tests for correctness
├── LICENSE # MIT License
└── README.md # This document
Although Match does not have any dependencies, the demos do. Demos import matplotlib, but you can skip (or comment) plotting cells (code) and not miss out on much. Demos come in two flavors: Jupyter Notebooks and Python scripts. These files are synced using Jupytext.
demo_linear_1d.mp4
The library uses implements reverse-mode automatic differentiation. Here are highlights of the implementation:
list2d.pycontains an implementation of several matrix operations (e.g., element-wise arithmetic, matrix multiplication, etc.)matrix.pyrelies onlist2d.pyand adds automatic differentiation functionality (it was important to decouple the underlying matrix operations inlist2d.pyfrom the usage ofMatrixobjects; this made it easier to include matrix operations in the gradient functions without running into a recursive loop)nn.pyadds common neural network functionality on top ofmatrix.py
You'll see some methods with a trailing _ in the name. This denotes that the method will mutate the current object. For example,
class List2D(object):
...
def ones_(self) -> None:
"""Modify all values in the matrix to be 1.0."""
self.__set(1.0)
...
def relu(self) -> List2D:
"""Return a new List2D object with the ReLU of each element."""
vals = [
[max(0.0, self.vals[i][j]) for j in range(self.ncol)]
for i in range(self.nrow)
]
return List2D(*self.shape, vals)
...The ones_ method sets all values of the current List2D to 1.0 whereas the relu method returns a new List2D object where each value is computed using relu function.
Some method names start with two _. These are internal functions that should not be used outside of the class. For example,
class List2D(object):
...
def __set(self, val) -> None:
"""Internal method to set all values in the matrix to val."""
self.vals = [[val] * self.ncol for _ in range(self.nrow)]
...You'll also find many double _ methods. These are known as Python magic or "dunder" (double underscore) methods. They make it easy to implement things like addition and subtraction with a custom object. For example,
class List2D(object):
...
def __add__(self, rhs: float | int | List2D) -> List2D:
"""Element-wise addition: self + rhs."""
return self.__binary_op(add, rhs)
...
a = List2D(...)
b = List2D(...)
c = a + b # <-- uses the __add__ method with self=a and rhs=bConsider the following model:
class MatchNetwork(match.nn.Module):
def __init__(self, n0, n1, n2) -> None:
super().__init__()
self.linear1 = match.nn.Linear(n0, n1)
self.relu = match.nn.ReLU()
self.linear2 = match.nn.Linear(n1, n2)
self.sigmoid = match.nn.Sigmoid()
def forward(self, x) -> Matrix:
x = self.linear1(x)
x = self.relu(x)
x = self.linear2(x)
x = self.sigmoid(x) # <-- this line of code is referenced below
return xWhat happens we we create a MatchNetwork and then compute an output?
num_features = 3
num_outputs = 1
model = MatchNetwork(n0=num_features, n1=4, n2=num_outputs)
x = ... # Some input example
y = ... # Some matching output label
yhat = model(x) # <-- invokes the forward passWe can represent the forward pass using math, or more readably with the following graph:
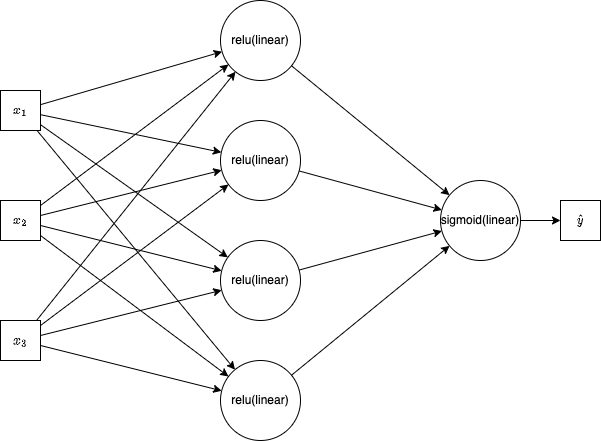
Although the figure above is useful, it is more useful (here) to think about the compute graph. But first, let's see the code needed to create the compute graph. Consider match's implementation of a sigmoid.
# Sigmoid object from nn.py
class Sigmoid(Module):
"""Sigmoid(x) = 1 / (1 + e^(-x))"""
def forward(self, x: Matrix) -> Matrix:
# Returns a new Matrix
return x.sigmoid()
# Sigmoid compute node from matrix.py
def sigmoid(self) -> Matrix:
"""Element-wise sigmoid."""
result = Matrix(self.data.sigmoid(), children=(self,))
def _gradient() -> None:
self.grad += result.data * (1 - result.data) * result.grad
result._gradient = _gradient
return result
# Sigmoid math from list2d.py
def sigmoid(self) -> List2D:
"""Return a new List2D object with the sigmoid of each element."""
vals = [
[sigmoid(self.vals[i][j]) for j in range(self.ncol)]
for i in range(self.nrow)
]
return List2D(*self.shape, vals)The following occurs when sigmoid(x) is called inside our model's forward method:
- We call
x.sigmoidwherexis the output of the previous linear node. - A new
Matrixobject (calledresult) is constructed- Elements in
resultare computed by taking thesigmoidof each value in the linear output - The the linear output becomes a child of
resultin the compute graph - The new matrix is the same shape as the linear output
- Elements in
- A gradient closure (called
_gradient) is created with the correct computations for future use- The closure is attached to the
resultcompute node - The closure has access to the values and gradient of the linear output
- The closure updates the gradient of the linear node (not itself)
- The closure is attached to the
Notice that we cannot compute the gradient of self until we have the gradient of result, which is not computed until its parents are computed. Here's the compute graph:
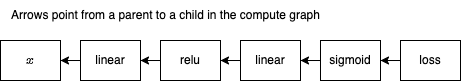
We now have a compute graph (it includes a reference to loss, which is given below). It was generated by the forward method of MatchNetwork, which was automatically called. Each method call in forward (self.linear1, etc.) builds up the graph. Now that we have the graph, we can compute an optimization function and update all model parameters.
loss = loss_fcn(yhat, y) # Any loss function will do
net.zero_grad() # Clear previous gradients (no-op first iteration)
loss.backward() # All gradients are computedFinally, we just need to update parameter values.
for param in net.parameters():
# param.grad was computed during the loss.backward() call above
param.data = param.data - learning_rate * param.gradSee 5.2 Backpropagation for more information.
To give you an idea of how to add some new functionality, let's trace through the process I used to add the mean-square-error loss function (MSELoss).
- Create a new class inside
nn.py:class MSELoss(Module)- The class must extend the
Moduleclass - Implement the
forwardmethod for the new class- It can take any arguments that fit your needs
- It should return a
Matrix - Most arguments will be a
Matrix(matrix in and matrix out, hence "forward") - For
MSELosswe return((target - prediction) ** 2).mean()
- The class must extend the
- Add necessary matrix operations to the
Matrixclass inmatrix.py. For MSE, we need- Subtraction for
(target - prediction)(implemented by defining the__sub__method) - Exponentiation for
_ ** 2(implemented by defining the__pow__method) - Mean for
_.mean()(implemented by defining themeanmethod) - You might also need to implement operators for the gradient computations (e.g.,
>for ReLU) - (note that each of these new methods must build up the compute graph)
- Subtraction for
- Add necessary math to the
List2Dclass inlist2d.py. For MSE, we need- (note that the
Matrixclass doesn't actually implement any math, just compute graph creation) - Subtraction is implemented by defining the
__sub__method - Exponentiation is implemented by defining the
__pow__method - Mean is implemented by defining the
meanmethod - (you may need to import new libraries or library functionality)
- (note that the
To recap:
- Create a new
Module - Add new functionality to
Matrixfor creating the compute graph - Add new functionality to
List2Dfor computing the matrix math
The library has a limited number of tests in the file test.py found in the root directory. Unit tests require the PyTorch library. They should be executed with:
# Run this command from the root of the repository (test.py should be in the root)
python -m unittest testThe sandbox directory can be ignored. That is where I am putting ideas for future updates.
- Build Your own Deep Learning Framework - A Hands-on Introduction to Automatic Differentiation - Part 2
- How we wrote xtensor 1/N: N-Dimensional Containers | by Johan Mabille
- Chain Rule on Matrix Multiplication
- A tiny scalar-valued autograd engine and a neural net library on top of it with PyTorch-like API
- The magic behind autodiff | Tutorials on automatic differentiation and JAX
- Google Colab: Coding a neural net
- Example matrix gradients
- Differentiable Programming from Scratch – Max Slater – Computer Graphics, Programming, and Math