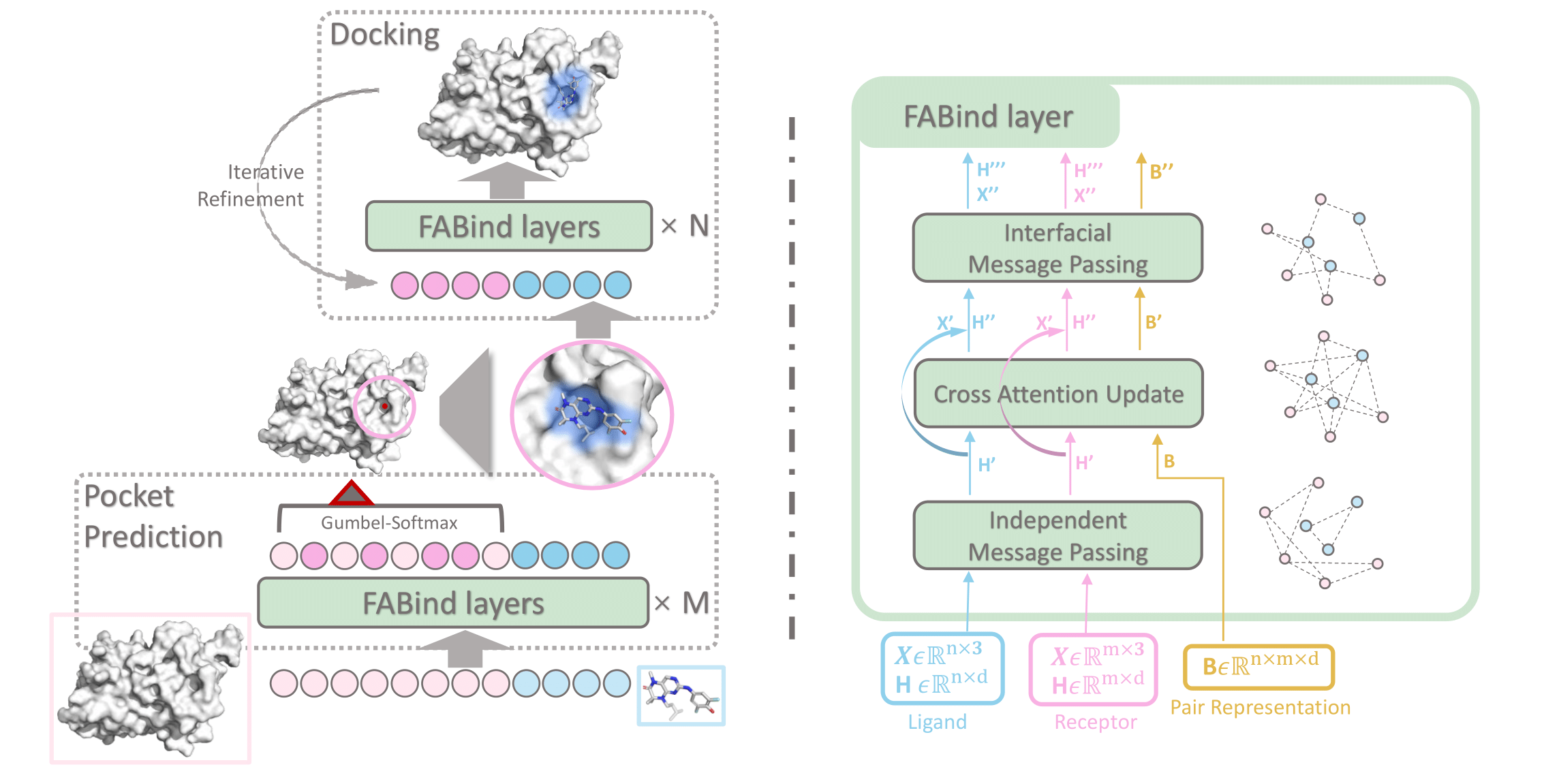
This repository contains the source code for NeurIPS 2023 paper "FABind: Fast and Accurate Protein-Ligand Binding". FABind achieves accurate docking performance with high speed compared to recent baselines. If you have questions, don't hesitate to open an issue or ask me via qizhipei@ruc.edu.cn, Kaiyuan Gao via im_kai@hust.edu.cn, or Lijun Wu via lijuwu@microsoft.com. We are happy to hear from you!
Jan 01 2024: Upload trained checkpoint into Google Drive.
Nov 09 2023: Move trained checkpoint from Github to HuggingFace.
Oct 10 2023: The trained FABind model and processed dataset are released!
Oct 11 2023: Initial commits. More codes, pre-trained model, and data are coming soon.
This is an example of how to set up a working conda environment to run the code. In this example, we have cuda version==11.3, and we install torch==1.12.0. To make sure the pyg packages are installed correctely, we directly install them from whl.
As the trained model checkpoint is included in the HuggingFace repository with git-lfs, you need to install git-lfs to pull the data correctly.
sudo apt-get install git-lfs # run this if you have not installed git-lfs
git lfs install
git clone https://github.com/QizhiPei/FABind.git --recursive
conda create --name fabind python=3.8
conda activate fabind
conda install pytorch==1.12.0 torchvision==0.13.0 torchaudio==0.12.0 cudatoolkit=11.3 -c pytorch
pip install https://data.pyg.org/whl/torch-1.12.0%2Bcu113/torch_cluster-1.6.0%2Bpt112cu113-cp38-cp38-linux_x86_64.whl
pip install https://data.pyg.org/whl/torch-1.12.0%2Bcu113/torch_scatter-2.1.0%2Bpt112cu113-cp38-cp38-linux_x86_64.whl
pip install https://data.pyg.org/whl/torch-1.12.0%2Bcu113/torch_sparse-0.6.15%2Bpt112cu113-cp38-cp38-linux_x86_64.whl
pip install https://data.pyg.org/whl/torch-1.12.0%2Bcu113/torch_spline_conv-1.2.1%2Bpt112cu113-cp38-cp38-linux_x86_64.whl
pip install https://data.pyg.org/whl/torch-1.12.0%2Bcu113/pyg_lib-0.2.0%2Bpt112cu113-cp38-cp38-linux_x86_64.whl
pip install torch-geometric
pip install torchdrug==0.1.2 rdkit torchmetrics==0.10.2 tqdm mlcrate pyarrow accelerate Bio lmdb fair-esm tensorboard
pip install fair-esmThe PDBbind 2020 dataset can be download from http://www.pdbbind.org.cn. We then follow the same data processing as TankBind.
We also provided processed dataset on zenodo. If you want to train FABind from scratch, or reproduce the FABind results, you can:
- download dataset from zenodo
- unzip the
zipfile and place it intodata_pathsuch thatdata_path=pdbbind2020
Before training or evaluation, you need to first generate the ESM2 embeddings for the proteins based on the preprocessed data above.
data_path=pdbbind2020
python fabind/tools/generate_esm2_t33.py ${data_path}Then the ESM2 embedings will be saved at ${data_path}/dataset/processed/esm2_t33_650M_UR50D.lmdb.
The pre-trained model is placed at ckpt/best_model.bin, which will be automatically downloaded when cloning this reporsitory with --recursive.
You can also manually download the pre-trained model from Hugging Face or Google Drive.
data_path=pdbbind2020
ckpt_path=ckpt/best_model.bin
python fabind/test_fabind.py \
--batch_size 4 \
--data-path $data_path \
--resultFolder ./results \
--exp-name test_exp \
--ckpt $ckpt_path \
--local-evalHere are the scripts available for inference with smiles and according pdb files.
The following script iteratively runs:
- Given smiles in
index_csv, preprocess molecules withnum_threadsmultiprocessing and save each processed molecule to{save_pt_dir}/mol. - Given protein pdb files in
pdb_file_dir, preprocess protein information and save it to{save_pt_dir}/processed_protein.pt. - Load model checkpoint in
ckpt_path, save the predicted molecule conformation inoutput_dir. Another csv file inoutput_dirindicates the smiles and according filename.
index_csv=../inference_examples/example.csv
pdb_file_dir=../inference_examples/pdb_files
num_threads=1
save_pt_dir=../inference_examples/temp_files
save_mols_dir=${save_pt_dir}/mol
ckpt_path=../ckpt/best_model.bin
output_dir=../inference_examples/inference_output
cd fabind
echo "====== preprocess molecules ======"
python inference_preprocess_mol_confs.py --index_csv ${index_csv} --save_mols_dir ${save_mols_dir} --num_threads ${num_threads}
echo "====== preprocess proteins ======"
python inference_preprocess_protein.py --pdb_file_dir ${pdb_file_dir} --save_pt_dir ${save_pt_dir}
echo "====== inference begins ======"
python fabind_inference.py \
--ckpt ${ckpt_path} \
--batch_size 4 \
--seed 128 \
--test-gumbel-soft \
--redocking \
--post-optim \
--write-mol-to-file \
--sdf-output-path-post-optim ${output_dir} \
--index-csv ${index_csv} \
--preprocess-dir ${save_pt_dir}data_path=pdbbind_2020
# write the default accelerate settings
python -c "from accelerate.utils import write_basic_config; write_basic_config(mixed_precision='no')"
# "accelerate launch" will run the experiments in multi-gpu if applicable
accelerate launch fabind/main_fabind.py \
--batch_size 3 \
-d 0 \
-m 5 \
--data-path $data_path \
--label baseline \
--addNoise 5 \
--resultFolder ./results \
--use-compound-com-cls \
--total-epochs 500 \
--exp-name train_tmp \
--coord-loss-weight 1.0 \
--pair-distance-loss-weight 1.0 \
--pair-distance-distill-loss-weight 1.0 \
--pocket-cls-loss-weight 1.0 \
--pocket-distance-loss-weight 0.05 \
--lr 5e-05 --lr-scheduler poly_decay \
--distmap-pred mlp \
--hidden-size 512 --pocket-pred-hidden-size 128 \
--n-iter 8 --mean-layers 4 \
--refine refine_coord \
--coordinate-scale 5 \
--geometry-reg-step-size 0.001 \
--rm-layernorm --add-attn-pair-bias --explicit-pair-embed --add-cross-attn-layer \
--noise-for-predicted-pocket 0 \
--clip-grad \
--random-n-iter \
--pocket-idx-no-noise \
--pocket-cls-loss-func bce \
--use-esm2-feat@article{pei2023fabind,
title={FABind: Fast and Accurate Protein-Ligand Binding},
author={Pei, Qizhi and Gao, Kaiyuan and Wu, Lijun and Zhu, Jinhua and Xia, Yingce and Xie, Shufang and Qin, Tao and He, Kun and Liu, Tie-Yan and Yan, Rui},
journal={arXiv preprint arXiv:2310.06763},
year={2023}
}
@inproceedings{
pei2023fabind,
title={{FAB}ind: Fast and Accurate Protein-Ligand Binding},
author={Qizhi Pei and Kaiyuan Gao and Lijun Wu and Jinhua Zhu and Yingce Xia and Shufang Xie and Tao Qin and Kun He and Tie-Yan Liu and Rui Yan},
booktitle={Thirty-seventh Conference on Neural Information Processing Systems},
year={2023},
url={https://openreview.net/forum?id=PnWakgg1RL}
}
We appreciate EquiBind, TankBind, E3Bind, DiffDock and other related works for their open-sourced contributions.