The project constitutes of two main subprojects (reflected in the subparsers), namely:
Corpusextraction and text normalization,- Preprocessing,
Clustering & Classification; along with the evaluations of the models and respective visualizations.
Corpus has been extracted from wikidata using Sparql. The respective queries provide links to 6 groups of interest - painters, writers, singers, politicians, architects, mathematicians wchich constitute 2 bigger clusters - 3 former - artists, 3 latter - non-artists.
Wikidata's API and its Python's wrapper - wptools take a while to query and process the obtained data; hence the wiki-clustering by default caches the normalized results by saving data and its corresponding schema after the extraction. This allows one to extract as many corpora as desired; the sizes can be adjusted by specifying respective attributes for the extraction as parameters to the script. By initially preparing several corpora one could later focus solely on investigating different settings and tuning hyper-parameters to find the most promising setup for the task of interest.
Before saving, the data is normalized - tokenization, lowercasing, and filtering out meaningless wordforms (function words) not to pollute the data. Since the goal doesn't regard understanding the structure of the data, leaving those tokens out in the corpus would only complicate the learning process. Additionally, we remove all the punctuation.
Initial representation is further processed to extract simple NLP-based features such as extraction of tokens matching certain part of speech in their base form (via lemmatization), and detecting named entities. Based on the outputs, results are being stored as separate attributes respectively. The prefix of the column is left from the original column, whilst the suffix contains the information about what has been extracted / processed. It is up to the user to decide which subset of attributes would be useful for the task of interest. It has been observed that among the simple setups the prominent results can be obtained using just tokenized content attribute. This may vary, e.g. depending on the size of the corpus.
After the whole process, the script creates vocabulary and dictionaries mapping tokens to their corresponding ids and back to their natural language (NL) representation. The script allows toggling between the two representations back and forth (from textual data to numeric and vice versa). The numerical representation is desired format by any machine learning model; whereas NL-form is better understood by humans. The latter could have been used, for instance, to preview the models' predictions in a human-readable manner.
Wiki-clustering implements both clustering and classification. Both methods can be customized in terms of prediction type; we distinguish between group-level clustering / classification and - category-level. The former one tries to identify data points as artists vs. non-artists; whereas the latter one predicts persons' occupation category painters vs. writers vs. singers vs. politicians vs. architects vs. mathematicians. For the clustering algorithm, we've utilized scikit learn's KMeans, whereas for the classification task we've implemented a custom PyTorch model of logistic regression with a single hidden and an output layers. After the forward pass results are being passed through a softmax activation layer to obtain predictions' estimates per group or category depending on the used approach. Weights in the hidden layer are of the N_MINIBATCH x N_HIDDEN shape, whereas output layer's are - N_HIDDEN x N_OUT_CLASSES resulting in the output of shape N_MINIBATCH x N_OUT_CLASSES. This behaviour is desired since we're interested in getting a prediction of the class per each data point in the dataset (in this mini-learning case given by a data mini-batch).
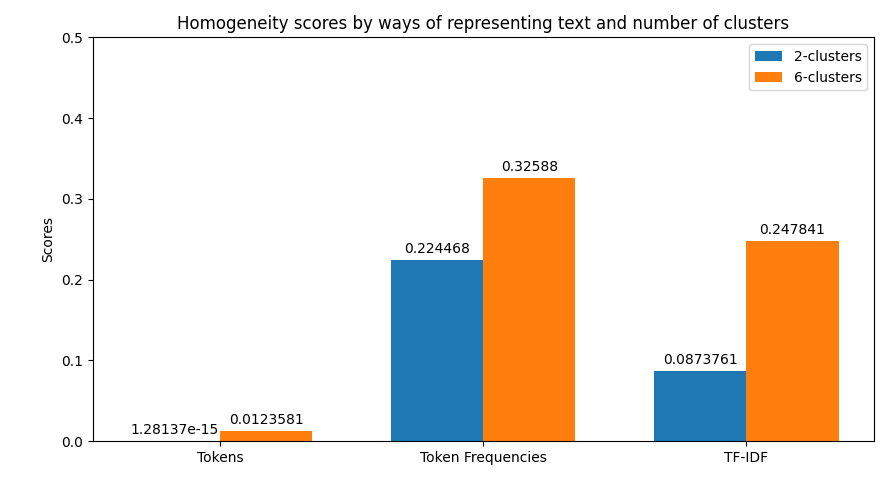
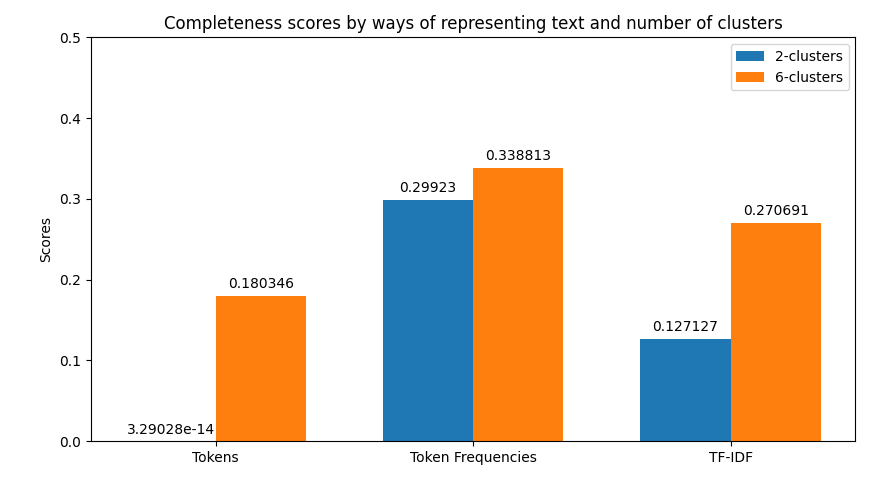
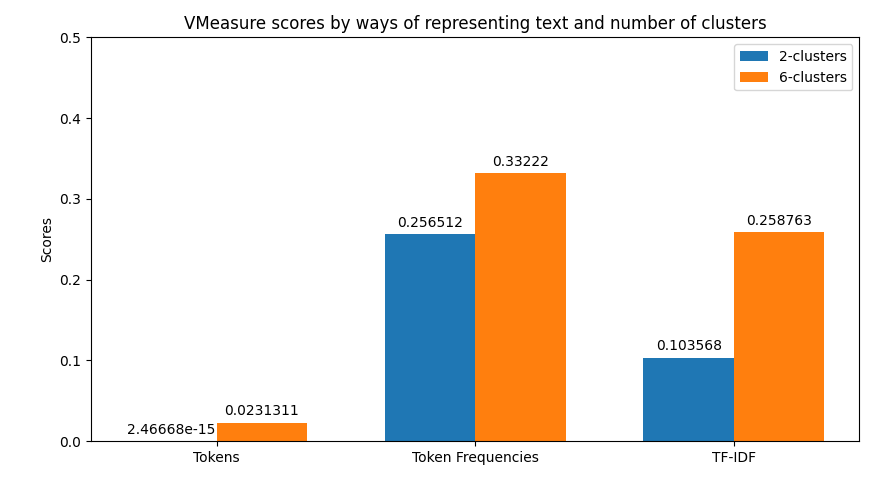
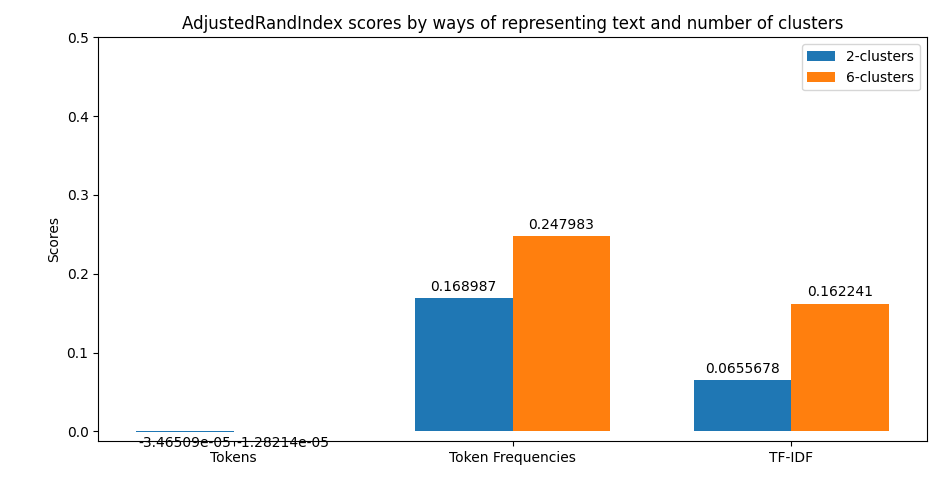
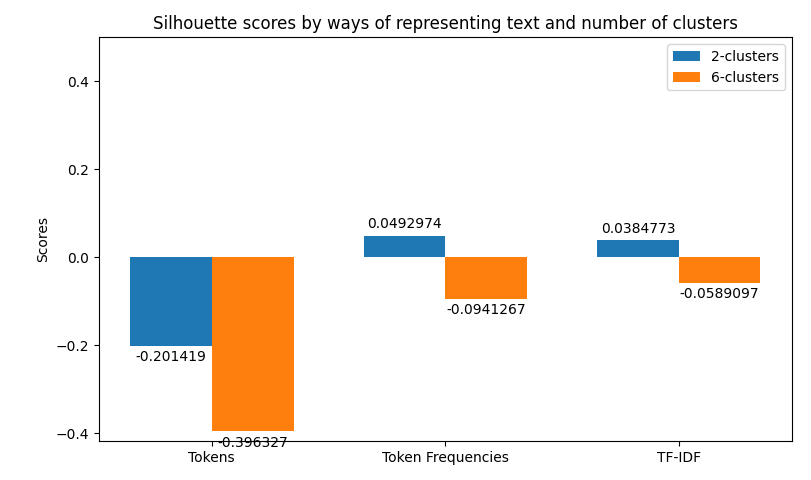
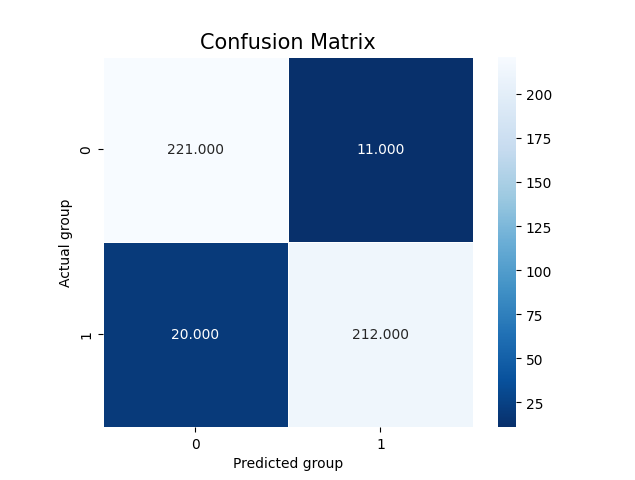
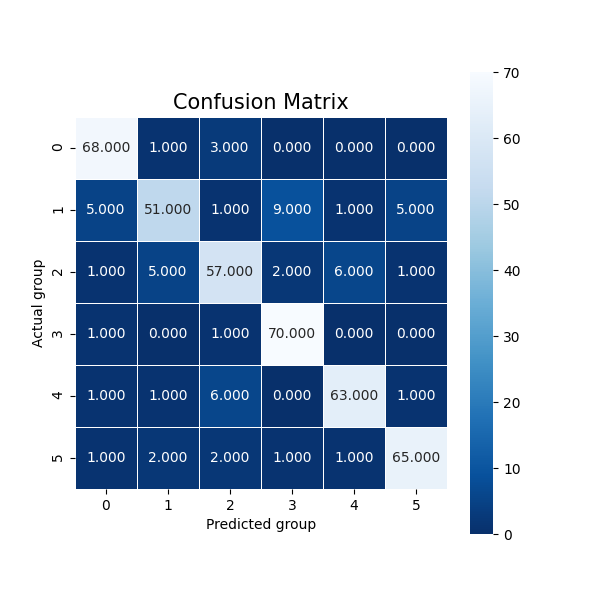
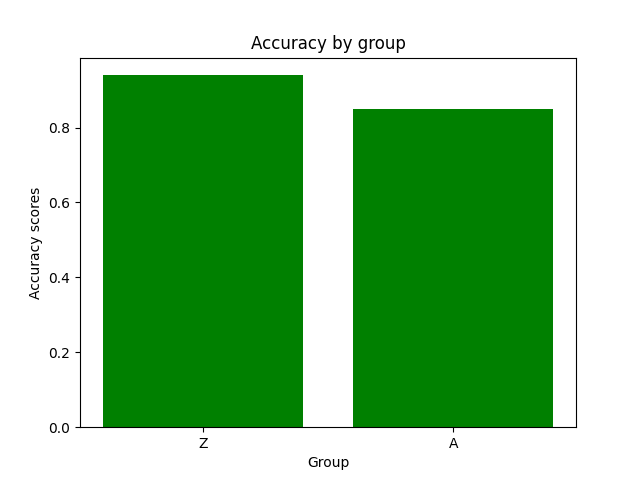
In the case of clustering evaluation of the model is given by both intrinsic and extrinsic methods, namely: Homegoeneity, Completeness, V Measure, Adjusted Rand Index, and Silhouette scores. Each evaluation is visualized as a juxtaposition between clusters of size 2 (for groups) and 6 (for categories) giving a comparison between the two. In the case of classification, we're allowed to perform a more robust evaluation since we have the golden dataset to measure the predictions against. For this reason, we're computing confusion matrices of each model's evaluation (resulting in two figures - one per class setting). Additionally, we're computing precision, recall, f1 score, and accuracy. The first four ones have been computed in the macro-scale; whereas the last one is simply calculated as the average of accuracies between different classes. Accuracy scores have been used as a metric to be visualized but visualizing method is flexible enough to easily be adjusted to visualize other metrics if only accumulated results of testing are stored in the correct format.
Move into the desired location in your local file system and execute the following command:
$ git clone https://github.com/amillert/wiki-clustering.gitOnce it's done, all the required files are at your disposal.
The environment.yml file contains all the information to successfully recreate the anaconda virtual environment along with the correct python version and all the compatible versions of libraries used in the project. To build the virtual environment, make sure anaconda is available in your system by typing:
$ conda --versionIn case, it's not available refer to the official manual at https://docs.anaconda.com/anaconda/install/.
If anaconda tool is at your disposal, one can proceed and recreate the virtual environment. Simply run the command:
$ conda env create -f environment.ymlThe virtual environment's name is amill_nakaz_2021 and it can be activated by typing:
$ conda activate amill_nakaz_2021In order to deactivate the anaconda virtual environment, one can run the command:
$ conda deactivateAs mentioned before, the main project constitutes two subprojects - corpus, and prediction. The distinction between the two and their respective arguments is reflected in the utils/argparser.py file. The two different subprojects are related to each other but should generally be treated as separate. Each of them comes with a set of parameters guiding the logic of execution and configuration.
| Id | Full parameter | Alternative | Functionality |
|---|---|---|---|
| 1 | --num_entries |
-n |
Specifies the amount of persons to be extracted per occupation |
| 2 | --sentences_per_article |
-k |
Specifies the amount of sentences to be extracted per entry |
| 3 | --parallel |
-p |
Flag which specify whether extraction should be run in parallel |
| 4 | --save_path |
-s |
Specifies a path to save the corpus |
Nota bene:
- If there are not enough sentences per entry, it will be discarded.
- The level of parallelization (without enforcing some messaging mechanism between actors) is limited by the fact that we need to extract
--num_entiresper category. Additionally we don't have a set of persons at our disposition beforehand since it's being extracted at the runtime as well so we can't create a pool of threads which continuously take new jobs after they're done with executing a small subtask. Because of all that, we decided to parallelize the job of extracting, filtering, collecting, and processing simply per each category. So in the busiest time of execution, there are only 6 processes running asynchronously. - The corpus is being stored as a
*.tsvfile along with the corresponding schema allowing to load data in the correct format (list object cannot be pickled). The output path should be provided as a directory; in the other case, data path should at least be in the correct format:corpus_<NUMBER>.tsv. Once, the saved file's path format has been slightly changed, and the suffix number isn't the next in the sequence, the automatic loading from provided directory will not work since the suffix number may not be found among the saved files.
| Id | Full parameter | Alternative | Functionality |
|---|---|---|---|
| 1 | --saved_path |
-s |
Specifies the path to saved corpus (to load) |
| 2 | --keep_top_tokens |
-k |
Specifies how many of the top-scored tokens should be kept in TFIDF |
| 3 | --batch_size |
-b |
Specifies the size of mini-batch for classification |
| 4 | --epochs |
-x |
Specifies the amount of epochs for classification |
| 5 | --eta |
-t |
Specifies the learning rate for classification |
| 6 | --n_hidden |
-h |
Specifies the size of hidden layer for classification |
Nota bene:
--saved_pathparameter can take either directory path leading to saved corpora or the exact file's path. The former will work only if all the files have been saved automatically by providing a directory path while saving corpora incorpussubproject. In the case, some custom file path has been provided, automatic data loading from the directory will not work and the user must provide this exact file's path. For this very reason, it's highly advised that a user provides a directory path in bothcorpusandpredictionsubprojects.- TFIDF in clustering is performed per each attribute; the results are later combined by stacking the sparse arrays back together.
- Parameters
--batch_size,--epochs,--eta,--n_hiddenspecify respective hyperparameters for classification; names should be self-explanatory.
We will start of by activating a conda environment:
$ conda activate amill_nakaz_2021Once, it's done we can run the script; we're interested in obtaining the corpus consisting of 30 persons per category with 10 sentences of content per entry. We specify the parallelization flag:
python main.py corpus -n 30 -k 10 -s data/raw-corpus -pThe resulting files will be located under wiki-clustering/data/raw-corpus/ as corpus_0.tsv and schema_0.json. The files' structure can be previewed in the following listings. First regards the corpus file:
title description content category group
Claude_Monet ['french', 'impressionist', 'painter', ...] painters A
Diego_Velázquez ['spanish', 'painter', ...] painters AThe second is its schema:
{
"title": "str",
"description": "list",
"content": "list",
"category": "str",
"group": "str"
}Having corpus ready, we can move on to clustering and classification. We can run both by choosing the second subparser with the following parameters:
python main.py prediction -b 16 -x 10 -t 0.8 -k 0.35 -s data/raw-corpus -d 10The first thing provided to the user is a visualization of the metrics retrieved from clustering. Their examples can be previewed below.
In the later part of the execution - classification, we will obtain visualization of the confusion matrices:
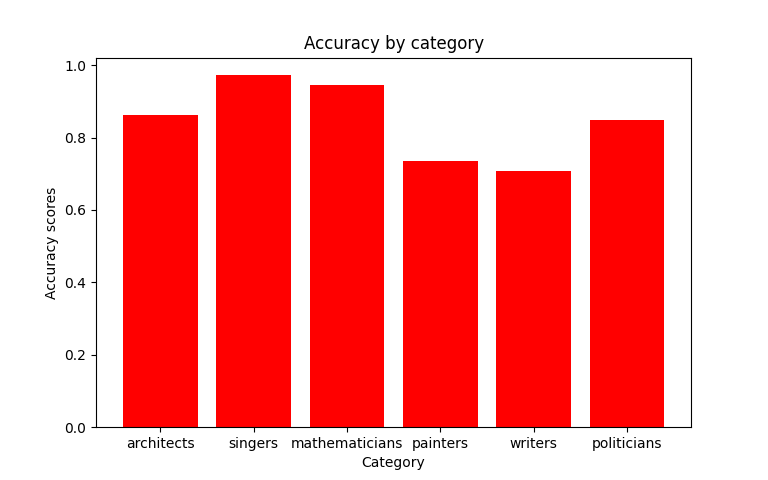
Which are shortly followed by the juxtaposition of averaged accuracies obtained for each class (whether it's a group or category as previously described).
-
Exercise 1-1 The SPARQL's queries used to automatically retrieve the names for 6 categories can be found under:
utils/__init__.py. -
Exercise 1-2, 1-3 The logic behind extracting entries from
Wikidataand then initial text normalization can be found under:handle-wiki/extraction.py. -
Exercise 2 The core data preprocessing can be found under:
corpus/preprocessing.py; however, the structure of the data is specified inutils/__init__.py. -
Exercise 3 The clustering process can be found under:
prediction/run.py, visualization is located in:utils/visualization.py. -
Exercise 4 The classification process can be found under:
prediction/run.py, visualization is located in:utils/visualization.py. The classifier model is declared inprediction/model/classifier.py. -
Corpus Expected corpus extracted for 30 persons per category with 10 sentences of content each can be found under
data/raw-corpus/corpus_0.tsv. Using this file (and schema belonging to it), one can simply proceed with the clustering & classification process.