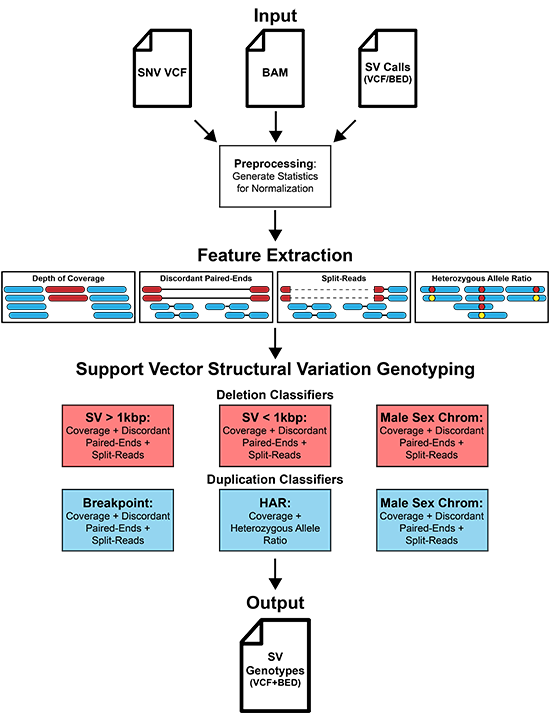
Support Vector Structural Variation Genotyper
A genotyper for the rest of us
Danny Antaki, William M Brandler, Jonathan Sebat; SV2: Accurate Structural Variation Genotyping and De Novo Mutation Detection from Whole Genomes, Bioinformatics, , btx813, https://doi.org/10.1093/bioinformatics/btx813
SV2 filters and integrates structural variants from multiple calling algorithms. Given multiple samples, SV2 creates a genotype matrix. SV2 also provides annotations for genes, repeat elements, and common SVs for filtering post-genotyping.
- User Guide
- Getting Started
- Input
- Output
- Training
- Performance
- Requirements
- Citing SV2
- Troubleshooting
- License
- Contact
SV2 (support-vector structural-variant genotyper) is a machine learning algorithm for genotyping deletions and duplications from paired-end whole genome sequencing data. SV2 can rapidly integrate variant calls from multiple SV discovery algorithms into a unified callset with high genotyping accuracy and detection of de novo mutations.
1️⃣ Installation
Install with pip Recommended
$ pip install sv2
2️⃣ Configure SV2
# download required resource files
$ sv2 -download
# define fasta locations
$ sv2 -hg19 /full/path/to/hg19.fa [-hg38 hg38.fa -mm10 mm10.fa]
3️⃣ Run SV2
$ sv2 -i in.bam -v sv.vcf -snv in.vcf.gz -p in.ped
SV2 requires BAM/CRAM files, SVs to genotype, SNV VCF files, and PED files.
$sv2 -i <in.bam ...> -v <sv.vcf ...> -b <sv.bed ...> -snv <snv.vcf.gz ...> -p <fam.ped ...>
Documentation for required inputs:
Output is generated in the current working directory by default.
$ sv2 ... -O <output path prefix>
-
sv2_preprocessing/contains preprocessing output -
sv2_features/feature extraction output -
sv2_genotypes/genotype output
Output VCF contains annotations for genes and filters
-
SV2 can merge divergent breakpoints. By default this option is off.
Advanced users can retrain SV2 genotyping classifiers with the original or custom training set.
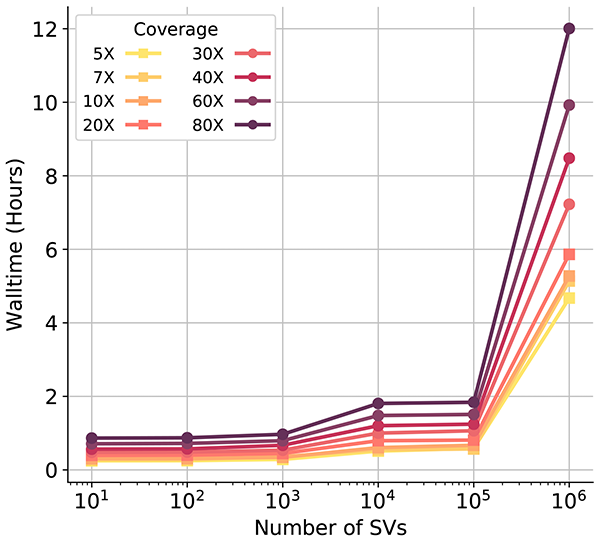
SV2 estimated runtime performance. Details
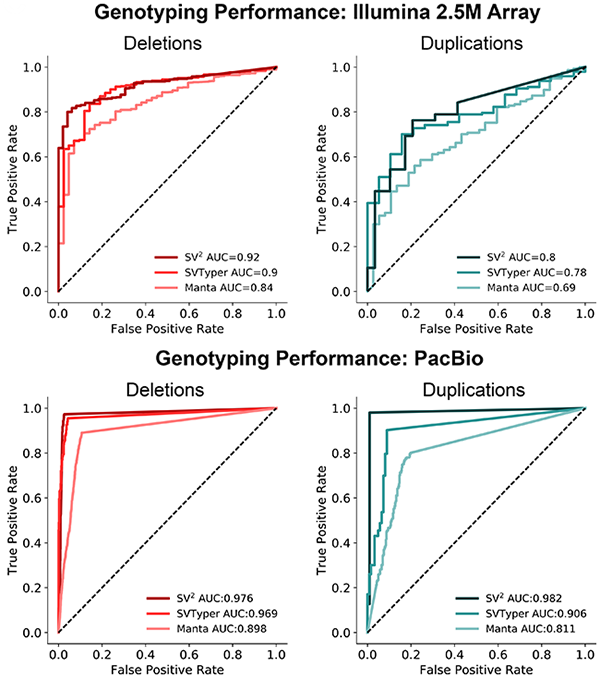
SV2 performance was determined with independent evaluation cohorts. Genotypes were validated with Illumina 2.5M microarrays and PacBio single molecule sequencing.
Please refer to the publication for performance details.
Danny Antaki, William M Brandler, Jonathan Sebat; SV2: Accurate Structural Variation Genotyping and De Novo Mutation Detection from Whole Genomes, Bioinformatics, , btx813, https://doi.org/10.1093/bioinformatics/btx813
MIT License
Copyright (c) 2017 Danny Antaki
Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the "Software"), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.
📫 dantaki@ucsd.edu 🤘