All data, results and code related to the paper can be found herewithin. For results that represent as large sets of images or tables, we provide scripts that are self-contained within this repository (i.e., all you need to do is to run them to reproduce our images) that allow you to reproduce these.
We have compiled a shiny app that allows you to explore the data and results interactively from your own browser, to see how you deploy and orient this tool go to the section "Shiny App" below.
A lot of people have inquired about access to the data used in this manuscript, all data is accessible at this Zenodo repository. However, the data is encrypted using 7z. To decrypt these files, use the following passwords:
- count matrices and images:
zNLXkYk3Q9znUseS - meta data and spot selection:
yUx44SzG6NdB32gY
For further questions, please see contact details below.
data/ST-cnts/: contains data for the 36 breast cancer sections used in this studyST-imgs/: contains the associated HE-images for the 36 sections used in this studyST-spotfiles/: contains tables with selected spots under tissue used to subset the raw gene count matricesgo-sets/: GO gene sets. Named go-{GO-ID}.tsvpublic.yaml: yaml file with links to the publicly available data sets that we've used
res/ST-pat/img/: images showing the pathologist's annotationslbl/: meta files (tsv) where each spot is labeled according to the pathologist's annotations
ST-cluster/lbl/: meta files (tsv) where each spot is labeled by membership of the expression based clustersmarkers/: marker genes for each clusterfea/: functional enrichment analysis results for each clusterpat-enr/: enrichment of clusters within the pathologist's regionscore-sig/: core signature for tumor and immune populationsmotivation.xlsx: motivations regarding how the clusters were annotated
ST-deconv/props/{major,minor,subset}.zip: spot-wise proportion estimates for respective tier, obtained using stereoscopepat-enr/{major,minor,subset}.zip: enrichment of cell types within the pathologist's regionscluster-enr/{major,minor,subset}.zip: enrichment of cell types within the expression based clusterscorrs/{major,minor,subset}.zip: correlation plots for all cell types within respective tier
ST-SW-enr/raw/: spot-wise pathway enrichment; values are given for within spot each sectionviz/: visualization of the raw spot-wise pathway enrichment
TLS-pred/coef-full.tsv: coefficient values (including intercept) for all genes in the model to predict TLS-scoretls-signature.tsv: the tls-signature presented in the papertls-fea.tsv: functional enrichment results for the tls-signature
scripts/-
Seurat_analysis_HER2_BC_samples.Rmd: markdown file outlining the gene-expression based clustering and associated analysis. -
* corrplot.R: script to use correlation plot. Uses bootstrapping to estimate confidence. (Figure 3 and 4) -
* enriched-region.py: estimates enrichment of types in defined regions by a permuation test (Figure 3 and 4). Produces palette-plots. -
* spw-enr.py: calculates spot-wise enrichment of gene set using Fisher's exact test (Figure 4) -
* rank-plot.py: generates a rank-plot where genes to highlight can be provided by a text file or as an argument. -
* viz-tls-score.py: visualized, B-cell and T-cell proportions together with calculated TLS-score -
* tls.py: fits the TLS-model. Takes as input a set of count matices and associated proportion values for these. -
* fea.py: conduct functional enrichment analysis using g:Profiler for a set of provided genes. Generates tsv file with all pathways and an image. (Figure 5) -
* predict.py: predict TLS-score for a ST/Visium section. Takes count data for the section to apply the model to and a coefficient file (generated by tls.py) -
* proportion-highlight.py: plots proportion estimates with specific regions highlighted. Uses a design-file to customize the character of the plot (seefigure3-config.yamlbelow). Used to generate Figure subplots 3A and 3B. -
mac-t_cell-interaction.ipynb: notebook outlining the analysis of the interferon pathways in the presence of certain macrophage and t-cell subsets. Used to generate Supplementary Figures 15-17.
-
rsc/figure3-config.yaml: configuration file toscripts/proportion-highlight.pyused to generate parts of Figure 3A and B.
app/data/: pre-processed count matrices, cell type proportion tables and images used for shiny application
* := with CLI (command line interface). Do ./script.py -h to see the different options that can be used.
While all the data and results are available as raw files, we have also constructed a tool that allows you to interactively explore these. The tool will open up in your default browser, but it runs locally (i.e. you host all the files on your own computer). See below for instructions regarding how to setup and orient the tool.
Begin by cloning this repository; to do so open a terminal, then enter a directory where you'd like to clone the repository into (here MY_DIR) and then do:
cd MY_DIR
git clone https://github.com/almaan/her2st.git .Next, we'll make sure that all the necessary packages (e.g., shiny) are installed. We've prepared an installation script for you - this will not overwrite you current package versions - that should take care of everything. However, if you're missing some dependencies (C++ backends) you might have to do a manual install. To check for and install missing packages; from the folder that you cloned this repo into, do:
cd her2st/app
./install-packages.R
NOTE : Make sure install-packages.R have the proper permissions (e.g., by doing chmod +x install-packages.R on a Linux computer) before running it.
If everything went as expected you should see a message like:
----
Successful installs:
----
ggplot2
jpeg
Matrix
grid
magrittr
magick
zeallot
shiny
shinyWidgets
shinycssloaders
shinydashboard
RColorBrewer
viridis
dplyr
extrafont
----
Failed installs:
----
NONE
In case you have any failed installs, try to install these manually and troubleshoot what dependencies you might be missing.
Once all the packages are installed - we are ready to run the app!
To launch the app, do:
cd MY_DIR/her2st/app
./launch.RNOTE : Just as for install-packages.R make sure launch.R have the proper permissions before running it.
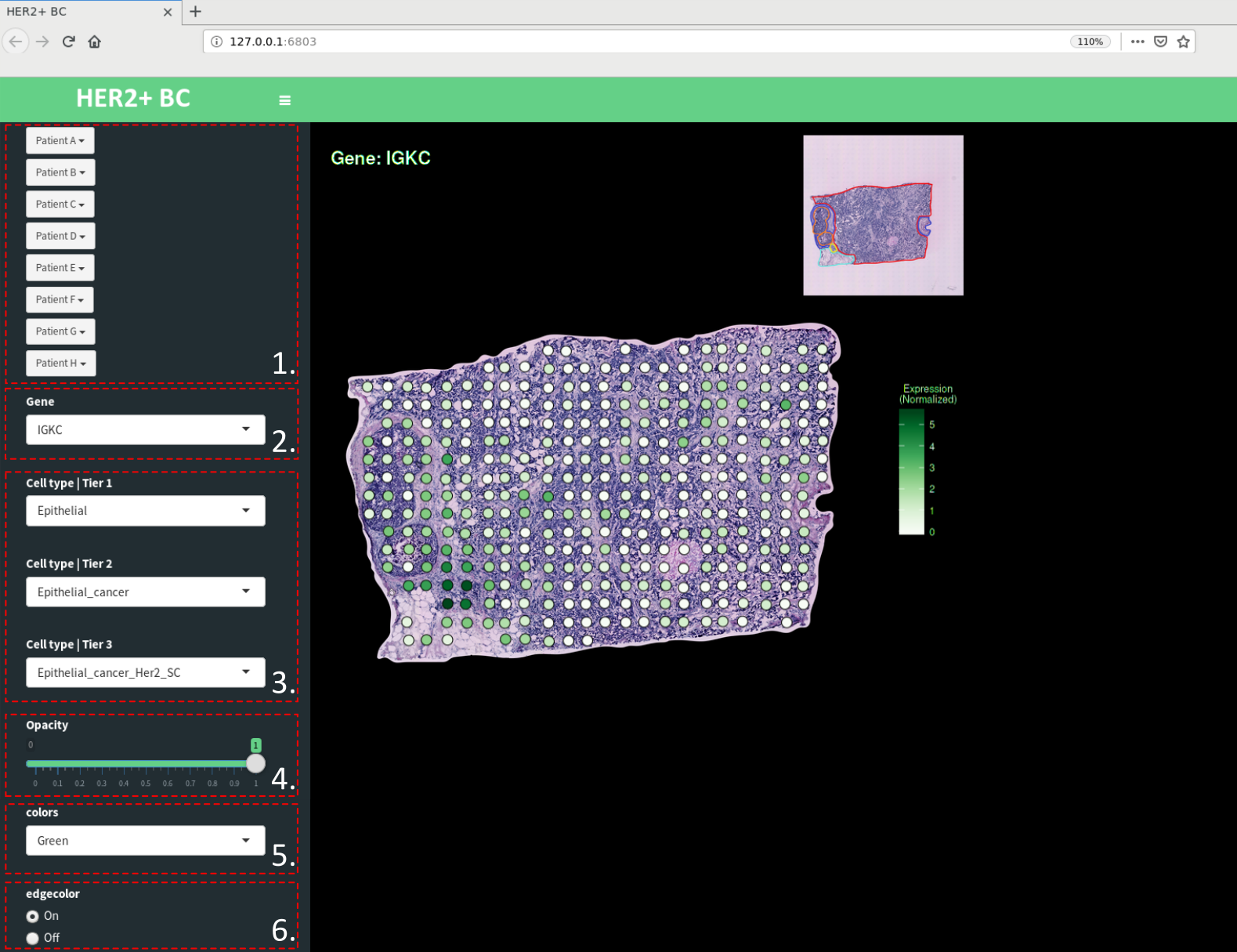
This should open a new tab or browser-window, with an address like 127.0.0.1:XXXX looking like:
As you can see you have some different options to choose from (indicated with dashed red boxes and a number). To elaborate some regarding these:
- Select section - click on the patient you want to visualize a section from, a drop-down menu will appear listing the different sections you can choose from.
- Gene - Select a gene of interest (GOI) to visualize the spatial expression of, overlaid on the tissue. You can either type the name your GOI or click the arrow to prompt a drop-down menu of genes to select from.
- Cell type - Select a tier and cell type (from a drop-down menu) to visualize the spatial distribution of this type (the proportion estimates) overlaid on the tissue.
- Opacity - Allows you to set the transparency of the markers
- Colors - Allows you to set the colormap used to visualize the values (default is Green)
- Edgecolor - Toggle whether edgecolor should be on or off
NOTE: Only one of the features in 2 and 3 can be visualized at a time, the last one selected will be displayed.
For questions related to the material presented at this site we recommend you to contact either:
- Alma Andersson : alma.andersson [at] differentiable [dot] net
- Ludvig Larsson : ludvig.larsson [at] scilifelab [dot] se
If you have code-specific questions, e.g., regarding the shiny app, opening an issue is the best way to communicate this if a quick response is desired.