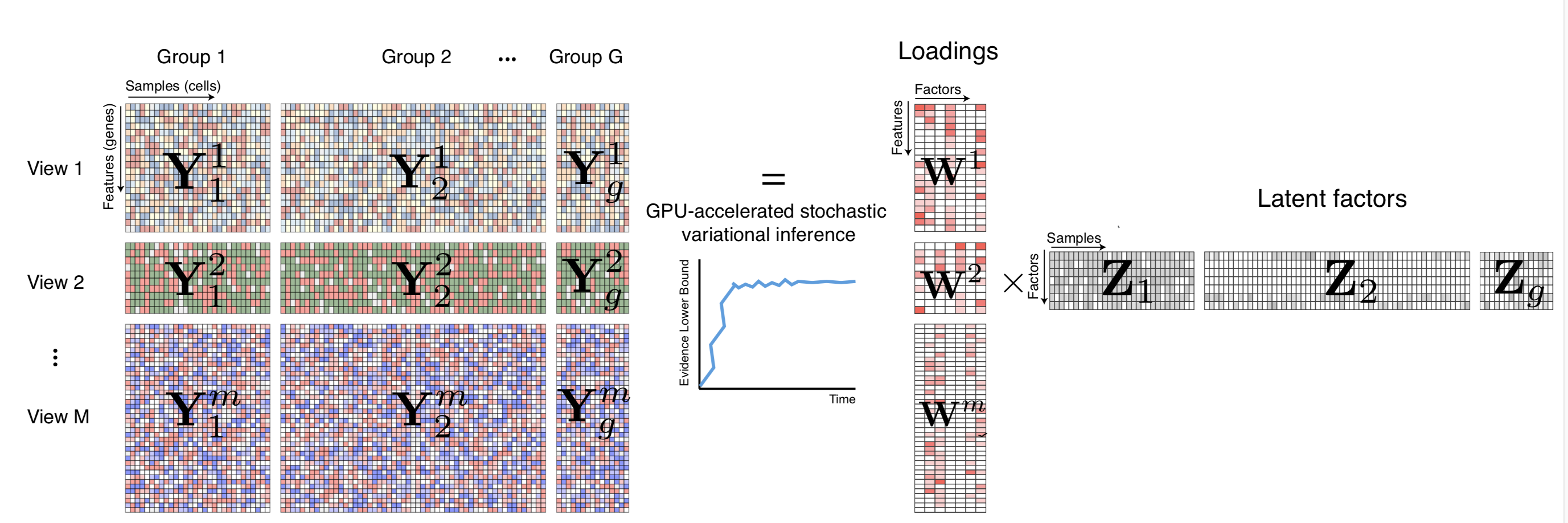
MOFA is a factor analysis model that provides a general framework for the integration of multi-omic data sets in an unsupervised fashion.
Intuitively, MOFA can be viewed as a versatile and statistically rigorous generalization of principal component analysis (PCA) to multi-omics data. Given several data matrices with measurements of multiple -omics data types on the same or on overlapping sets of samples, MOFA infers an interpretable low-dimensional data representation in terms of (hidden) factors. These learnt factors represent the driving sources of variation across data modalities, thus facilitating the identification of cellular states or disease subgroups.
In MOFA v2 (MOFA+) we added the following improvements:
- Multi-group functionality: intuitively, this breaks the assumption of independent samples and allows inference across multiple groups, where groups are predefined sets of samples (i.e. different conditions, batches, cohorts, etc.).
- Fast inference using a stochastic variational framework: this can be powered by GPUs: enabling inference with very large data sets.
For more details you can read our papers:
- MOFA (v1): http://msb.embopress.org/cgi/doi/10.15252/msb.20178124
- MOFA+: https://www.biorxiv.org/content/10.1101/837104v1
The core of MOFA is implemented in Python. However, the whole procedure can be run with R and we provide the downstream analysis functions only in R.
Python dependencies can be installed using pip (from the Unix terminal)
pip install mofapy2MOFA2 R package can be installed using R:
devtools::install_github("bioFAM/MOFA2/MOFA2", build_opts = c("--no-resave-data --no-build-vignettes"))You can build an image using the provided Dockerfile:
docker build -t mofa2 .
You will then be able to use R or Python from the container.
docker run -ti --rm -v $DATA_DIRECTORY:/data mofa2 R
# ^
# |
# use `-v` to map a folder on your machine to a container directory
The command above will launch R with MOFA2 and its dependencies installed while mounting $DATA_DIRECTORY to the container.
You can also pull the pre-build image from dockerhub.
- Getting started: general overview and description of the method.
- Training a model in R
- Training a model in Python (jupyter notebook)
- Downstream analysis (in R)
- Analysis of a multi-group scRNA-seq data set (in R): Figure 2 of the paper.
- Analysis of single-cell DNA methylation data (in R): Figure 3 of the paper, in preparation...
- Integration of single-cell multi-modal data (in R): Figure 4 of the paper.
- Using fast stochastic variational inference: in preparation...
- Analysis of CITE-seq data: in preparation...
- Analysis of chronic lymphocytic leukaemia cohort for personalised medicine: in preparation...
- Robustness analysis and model selection
The data and the pre-trained models can be downloaded here
We have a Slack group where we provide quick and personalised help, this is the link.
Ricard Argelaguet (ricard@ebi.ac.uk) and Danila Bredikhin (danila.bredikhin@embl.de)
(1.1) Can MOFA+ be applied to bulk data?
MOFA+ remains 100% applicable to bulk data.
(1.2) How does the multi-group inference work in MOFA+?
A group is simply defined as a predefined set of samples. There is total flexibility on how to define them, but they usually correspond to different conditions, cohorts, time points, etc. see our paper for details.
Very importantly, the groups are treates as batches of data, and the model is not focused on capturing differential changes between batches, but rather it is exploiting the coordinated variability between and within batches. Hence, prior to fitting the model the group effect is regressed out and features are centered to zero. This ensures that the model only looks at the variance, and not at the means.\
Technically, the multi-group inference is achieved by incorporating sparsity priors in the factors (see the supplementary methods of the paper).
(1.3) can I run MOFA v1 using MOFA+?
Yes, if you don't have multi-group structure in your data then just define a single group. This is equivalent to MOFA v1 (but significantly faster). However, due to some improvements in the parameter initialisation and the priors, you will not obtain identical results to your previous MOFA v1 models.
(1.4) Does MOFA+ inherit previous features from MOFA v1?
Yes, pretty much everything: handling of missing data, non-gaussian likelihoods and sparsity in the weights. The novel model features are additional sparsity priors in the factors and the improved inference scheme.
(2.1) How do I normalise the data?
Proper normalisation of the data is critical for the model to work. First, one needs to remove library size effects. For count-based data such as RNA-seq or ATAC-seq we recommend size factor normalisation + variance stabilisation. For microarray DNA methylation data, make sure that samples have no differences in the average intensity. If this is not done correctly, the model will learn a very strong Factor 1 that will capture this variability, and more subtle sources of variation will be harder to identify.
(2.2) Should I do any filtering to the input data?
It is strongly recommended that you filter highly variable features (HVGs) per assay.
Importantly, when doing multi-group inference, you have to regress out the group effect before selectinc HVGs. Otherwise you will enrich for features that show differences in the mean between groups.
(2.3) How many samples do I need?
Factor Analysis models are only useful with large sample sizes, let's say more than 15. With few samples you will detect very few relevant factors
(2.4) Should I remove undesired sources of variability (i.e. batch effects) before fitting the model?
Yes, if you have clear technical factors, we strongly encourage to regress it out a priori using a simple linear model. The reason for this is that the model will "focus" on the huge variability driven by the technical factors, and smaller sources of variability could be missed.
You can regress out known covaraites using the function regressCovariates. See the corresponding documentation and the CLL vignette for details.
(2.5) My data sets have different dimensionalities, does this matter?
Yes, this is important. Bigger data modalities will tend to be overrepresent in the MOFA model. It is good practice to filter features (based for example on variance, as lowly variable features provide little information) in order to have the different dimensionalities within the same order of magnitudes. If this is unavoidable, take into account that the model has the risk of missing (small) sources of variation unique to the small data set.
(2.6) Does MOFA handle missing values?
Yes! and there is no hidden imputation step, it simply ignores them. Matrix factorisation models are known to be very robust to the presence of missing values!
(3.1) I get one of the following errors when running MOFA:
AttributeError: 'module' object has no attribute 'core.entry_point
Error in py_module_import(module, convert = convert) :
ModuleNotFoundError: No module named 'mofapy2'
First thing: restart R and try again. If the error still holds, this means that either:
(1) you did not install the mofa Python package (see instructions above).
(2) you have multiple python installations and R is not detecting the correct one where mofa is installed. You need to find out the right Python interpreter, which usually will be the one you get when running which python in the terminal. You can test if the mofa packaged is installed by running INSIDE python: import mofapy2.
Once everything is figured out, specify the following at the beginning of your R script:
library(reticulate)
use_python("YOUR_PYTHON_PATH", required=TRUE)
You can also use use_conda instead of use_python if you work with conda environments. Read more about the reticulate package and how it integrates Python and R
(3.2) I get the following error when installing the R package:
ERROR: dependencies 'XXX', 'YYY' are not available for package 'MOFA2'
You probably tried to install them using install.packages(). These packages should be installed from Bioconductor.
(3.3) I hate R, can I do MOFA only with Python?
XXX
(4.1) How many factors should I learn?
Similar to other latent variable models, this is a hard question to answer. It depends on the data set and the aim of the analysis. If you want to get an overview on the major sources of variability then use a small number of factors (K<=10). If you want to capture small sources of variability, for example to do imputation or eQTL mapping, then go for a large number of factors (K>25).
(4.2) Can MOFA automatically learn the number of factors?
Yes, but the user needs to specify a minimum value of % variance explained. Then, MOFA will actively remove factors (during training) that explain less than the specified amount of variance.
If you have no idea on what to expect, it is better to start with a fixed number of factors and set the % variance threshold to 0.
(4.3) Can I include known covariates in the model?
We extensively tested this functionality and it was not yielding good results. The reason is that covariates are usually discrete labels that do not reflect the underlying molecular biology. For example, if you introduce age as a covariate, but the actual age is different from the “molecular age”, the model will simply learn a new factor that corresponds to this “latent” molecular age, and it will drop the covariate from the model.
We recommend that you learn the factors in a completely unsupervised manner and then relate them to the biological covariates (see vignettes). If your covariate of interest is an important driver of variability, do not worry, MOFA will find it!
(4.4) The weights have different values between runs. Is this expected?
This is normal and it happens because of two reasons. The first one is that the model does not always converge to the same exact solution (see below in the FAQ), although different model instances should be pretty similar. The second reason is that factor analysis models are rotation invariant. This means that you can rotate your factors and your weights and still find the same solution. This implies that the signs of the weight or the factors can NOT be compared across trials, only within a trial.
(4.5) What data modalities can MOFA cope with?
- Continuous data: modelled using a gaussian likelihood
- Binary data: modelled using a bernoulli likelihood
- Count data: using a poisson likelihood. Importantly, the use of non-gaussian likelihoods require further approximations and are not as accurate as the gaussian likelihood. Hence, if your data can be safely transformed to match the gaussian likelihood assumptions, this is ALWAYS recommended. For example RNA-seq data is expected to be normalised and modelled with a gaussian distribution, do not input the counts directly.
(4.6) The model does not converge smoothly, and it oscillates between positive and negative deltaELBO values
First, check that you are using the right likelihood model (see above). Second, make sure that you have no features or samples that are full of missing values. Third, check that you have no features with zero (or very little) variance. If the problem does not disappear, please contact us via mail or the Slack group.
(4.7) Does MOFA always converge to the same solutions?
No, as occurs in most complex Bayesian models, they are not guaranteed to always converge to the same solution.
In practice, however, we observed that the solutions are highly consistent, particularly for the top factors. However, we recommend doing a robustness analysis. This is done by training multiple model instances and check the correlation of the factors across the different solutions See the function compare_models().
(5.1) How can I do Gene Set Enrichment Analysis?
First, you need to create your binary gene set matrix where rows are feature sets and columns are features (genes). We have manually processed some of Reactome and MSigDB gene sets for mouse and human. Contact us if you would like to use the data.
Then, you will have to choose a local statistic per feature (the loading, by default), a global statistic per pathway (average loading, by default), and a statistical test. The most trustworthy one is a permutation test with a long number of iterations, but this is slow and a fast parametric tests is also available. However, note that it tends to inflate the p-values due to the correlation structure between related genes (see for example Gatti2010).