LucaProt(DeepProtFunc) is an open source project developed by Alibaba and licensed under the Apache License (Version 2.0).
This product contains various third-party components under other open source licenses.
See the NOTICE file for more information.
Notice:
This project provides the Python dependency environment installation file, installation commands, and the running command of the trained LucaProt model for inference or prediction, which can be found in this repository. These models are compatible with Linux, Mac OS, and Windows systems, supporting both CPU and GPU configurations for inference tasks.
LucaProt: A novel deep learning framework that incorporates protein amino acid sequence and structural information to predict protein function.
We developed a new deep learning model, namely, Deep Sequential and Structural Information Fusion Network for Proteins Function Prediction (DeepProtFunc/LucaProt), which takes into account protein sequence and structural information to facilitate the accurate annotation of protein function.
Here, we applied LucaProt to identify viral RdRP.
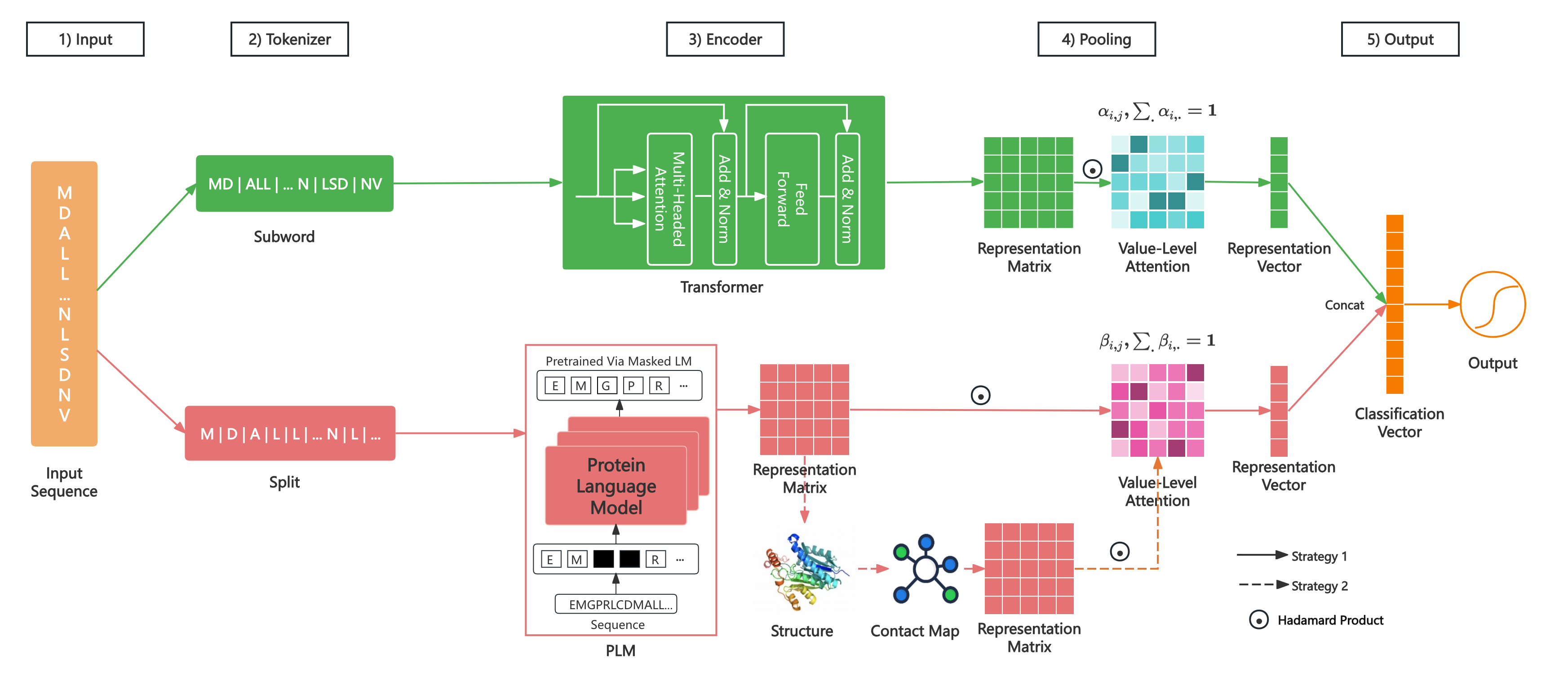
We treat protein function prediction as a classification problem. For example, viral RdRP identification is a binary-class classification task, and protein general function annotation is a multi-label classification task. The model includes five modules: Input, Tokenizer, Encoder, Pooling, and Output. Its architecture is shown in Figure 1.
Figure 1 The Architecture of LucaProt
Use the amino acid letter sequence as the input of our model. The model outputs the function label of the input protein, which is a single tag (binary-class classification or multi-class classification) or a set of tags (multi-label classification).
System: Ubuntu 20.04.5 LTS
Python: 3.9.13
Download anaconda: anaconda
Cuda: cuda11.7 (torch==1.13.1)
# Select 'YES' during installation for initializing the conda environment
sh Anaconda3-2022.10-Linux-x86_64.sh
# Source the environment
source ~/.bashrc
# Verification
conda
# Install env and python 3.9.13
conda create -n lucaprot python=3.9.13
# activate env
conda activate lucaprot
# Install git
sudo apt-get update
sudo apt install git-all
# Enter the project
cd LucaProt
# Install
pip install -r requirements.txt -i https://pypi.tuna.tsinghua.edu.cn/simple You can simply use this project to infer or predict for unknown sequences.
cd LucaProt/src/prediction/
sh run_predict_one_sample.sh
Note: the embedding matrix of the sample is real-time predictive.
Or:
cd LucaProt/src/
# using GPU(cuda=0)
export CUDA_VISIBLE_DEVICES="0,1,2,3"
python predict_one_sample.py \
--protein_id protein_1 \
--sequence MTTSTAFTGKTLMITGGTGSFGNTVLKHFVHTDLAEIRIFSRDEKKQDDMRHRLQEKSPELADKVRFFIGDVRNLQSVRDAMHGVDYIFHAAALKQVPSCEFFPMEAVRTNVLGTDNVLHAAIDEGVDRVVCLSTDKAAYPINAMGKSKAMMESIIYANARNGAGRTTICCTRYGNVMCSRGSVIPLFIDRIRKGEPLTVTDPNMTRFLMNLDEAVDLVQFAFEHANPGDLFIQKAPASTIGDLAEAVQEVFGRVGTQVIGTRHGEKLYETLMTCEERLRAEDMGDYFRVACDSRDLNYDKFVVNGEVTTMADEAYTSHNTSRLDVAGTVEKIKTAEYVQLALEGREYEAVQ \
--emb_dir ./emb/ \
--truncation_seq_length 4096 \
--dataset_name rdrp_40_extend \
--dataset_type protein \
--task_type binary_class \
--model_type sefn \
--time_str 20230201140320 \
--step 100000 \
--threshold 0.5 \
--gpu_id 0
# using CPU(gpu_id=-1)
python predict_one_sample.py \
--protein_id protein_1 \
--sequence MTTSTAFTGKTLMITGGTGSFGNTVLKHFVHTDLAEIRIFSRDEKKQDDMRHRLQEKSPELADKVRFFIGDVRNLQSVRDAMHGVDYIFHAAALKQVPSCEFFPMEAVRTNVLGTDNVLHAAIDEGVDRVVCLSTDKAAYPINAMGKSKAMMESIIYANARNGAGRTTICCTRYGNVMCSRGSVIPLFIDRIRKGEPLTVTDPNMTRFLMNLDEAVDLVQFAFEHANPGDLFIQKAPASTIGDLAEAVQEVFGRVGTQVIGTRHGEKLYETLMTCEERLRAEDMGDYFRVACDSRDLNYDKFVVNGEVTTMADEAYTSHNTSRLDVAGTVEKIKTAEYVQLALEGREYEAVQ \
--emb_dir ./emb/ \
--truncation_seq_length 4096 \
--dataset_name rdrp_40_extend \
--dataset_type protein \
--task_type binary_class \
--model_type sefn \
--time_str 20230201140320 \
--step 100000 \
--threshold 0.5 \
--gpu_id -1
-
--protein_id
str, the protein id. -
--sequence
str, the protein sequence. -
--truncation_seq_length
int, truncate sequences longer than the given value. Recommended values: 4096, 2048, 1984, 1792, 1534, 1280, 1152, 1024, default: 4096. -
--emb_dir(optional)
path, the saved dirpath of the protein predicted embedding matrix or vector during prediction, optional. -
--dataset_name
str, the dataset name for building of our trained model(rdrp_40_extend). -
--dataset_type
str, the dataset type for building of our trained model(protein). -
--task_type
str, the task type for building of our trained model(binary_class). -
--model_type
str, the model name for building of our trained model(sefn). -
--time_str
str, the running time string(yyyymmddHimiss) for building of our trained model(20230201140320). -
--step
int, the training global step of model finalization(100000). -
--threshold
float, sigmoid threshold for binary-class or multi-label classification, None for multi-class classification, default: 0.5. -
--gpu_id: int, the gpu id to use(-1 for cpu).
the samples are in *.fasta, sample by sample prediction.
-
--fasta_file
str, the samples fasta file. -
--save_file
str, file path, save the predicted results into the file. -
--print_per_number
int, print progress information for every number of samples completed, default: 100.
cd LucaProt/src/prediction/
sh run_predict_many_samples.shOr:
cd LucaProt/src/
# using GPU(cuda=0)
export CUDA_VISIBLE_DEVICES="0,1,2,3"
python predict_many_samples.py \
--fasta_file ../data/rdrp/test/test.fasta \
--save_file ../result/rdrp/test/test_result.csv \
--emb_dir ../emb/ \
--truncation_seq_length 4096 \
--dataset_name rdrp_40_extend \
--dataset_type protein \
--task_type binary_class \
--model_type sefn \
--time_str 20230201140320 \
--step 100000 \
--threshold 0.5 \
--print_per_number 10 \
--gpu_id 0
# using CPU(gpu_id=-1)
python predict_many_samples.py \
--fasta_file ../data/rdrp/test/test.fasta \
--save_file ../result/rdrp/test/test_result.csv \
--emb_dir ../emb/ \
--truncation_seq_length 4096 \
--dataset_name rdrp_40_extend \
--dataset_type protein \
--task_type binary_class \
--model_type sefn \
--time_str 20230201140320 \
--step 100000 \
--threshold 0.5 \
--print_per_number 10 \
--gpu_id -1The test data (small and real) is in demo.csv, where the 7th column of each line is the filename of the structural embedding information prepared in advance.
And the structural embedding files store in embs.
The test data includes 50 viral-RdRPs and 50 non-viral RdRPs.
cd LucaProt/src/prediction/
sh run_predict_from_file.sh
Or:
cd LucaProt/src/
# using GPU(cuda=0)
export CUDA_VISIBLE_DEVICES="0,1,2,3"
python predict.py \
--data_path ../data/rdrp/demo/demo.csv \
--emb_dir ../data/rdrp/demo/embs/esm2_t36_3B_UR50D \
--dataset_name rdrp_40_extend \
--dataset_type protein \
--task_type binary_class \
--model_type sefn \
--time_str 20230201140320 \
--step 100000 \
--evaluate \
--threshold 0.5 \
--batch_size 16 \
--print_per_batch 100 \
--gpu_id 0
# using CPU(gpu_id=-1)
python predict.py \
--data_path ../data/rdrp/demo/demo.csv \
--emb_dir ../data/rdrp/demo/embs/esm2_t36_3B_UR50D \
--dataset_name rdrp_40_extend \
--dataset_type protein \
--task_type binary_class \
--model_type sefn \
--time_str 20230201140320 \
--step 100000 \
--evaluate \
--threshold 0.5 \
--batch_size 16 \
--print_per_batch 100 \
--gpu_id -1
-
--data_path
path, the file path of prediction data, including 9 columns mentioned above. The value of Column Label can be null. -
--emb_dir
path, the saved dirpath of all sample's structural embedding information prepared in advance. -
--dataset_name
str, the dataset name for building of our trained model(rdrp_40_extend). -
--dataset_type
str, the dataset type for building of our trained model(protein). -
--task_type
str, the task name for building of our trained model(binary_class). -
--model_type
str, the model name for building of our trained model(sefn). -
--time_str
str, the running time string(yyyymmddHimiss) for building of our trained model(20230201140320). -
--step
int, the training global step of model finalization(100000). -
--threshold
float, sigmoid threshold for binary-class or multi-label classification, None for multi-class classification, default: 0.5. -
--evaluate(optional)
store_true, whether to evaluate the predicted results. -
--ground_truth_col_index(optional)
int, the ground truth col index of the ${data_path}, default: None. -
--batch size
int, batch size per GPU/CPU for evaluation, default: 16. -
--print_per_batch
int, how many batches are completed every time for printing progress information, default: 1000. -
gpu_id: int, the gpu id to use(-1 for cpu).
Note: the embedding matrices of all the proteins in this file need to prepare in advance($emb_dir).
11 verification datasets unrelated to the model building dataset, include 7 exists viral-RdRP datasets and 4 exists non viral-RdRP datasets.
Run the prediction python script https://github.com/alibaba/LucaProt/src/predict_many_samples.py
The performance on these 11 independent verification datasets of LucaProt.
LucaProt-Performance-On-11-Independent-Datasets.xlsx
or LucaProt Figshare
This project is used to predict unlabeled protein sequences and to measure the time spent.
LucaProtApp or LucaProt Figshare
LucaProt is suitably speedy because it only needs to predict the structural representation matrix rather than the complete 3D structure of the protein sequence.
Benchmark: For each sequence length range(total 10 groups), selected 50 viral-RdRPS and 50 non-viral RdRPs for each group for inference time cost calculation.
inference_time_data_of_github.csv
or LucaProt Figshare
Note: The spend time includes the time of the structural representation matrix inference, excludes the time of model loading.
Notice: when the sequence length does not exceed 1024, you can use the 24GB GPU for inference, such as the A10.
| Protein Seq Len Range | Average Time | Maximum Time | Minimum Time |
|---|---|---|---|
| 300 <= Len < 500 | 0.20s | 0.24s | 0.16s |
| 500 <= Len < 800 | 0.30s | 0.39s | 0.24s |
| 800 <= Len < 1,000 | 0.42s | 0.46s | 0.39s |
| 1,000 <= Len < 1,500 | 0.59s | 0.74s | 0.45s |
| 1,500 <= Len < 2,000 | 0.87s | 1.02s | 0.73s |
| 2,000 <= Len < 3,000 | 1.31s | 1.69s | 1.01s |
| 3,000 <= Len < 5,000 | 2.14s | 2.78s | 1.72s |
| 5,000 <= Len < 8,000 | 3.03s | 3.45s | 2.65s |
| 8,000 <= Len < 10,000 | 3.77s | 4.24s | 3.32s |
| 10,000 <= Len | 9.92s | 17.66s | 4.30s |
| Protein Seq Len Range | Average Time | Maximum Time | Minimum Time |
|---|---|---|---|
| 300 <= Len < 500 | 3.97s | 5.71s | 2.77s |
| 500 <= Len < 800 | 5.78s | 7.50s | 4.48s |
| 800 <= Len < 1,000 | 8.23s | 9.41s | 7.41s |
| 1,000 <= Len < 1,500 | 11.49s | 16.42s | 9.22s |
| 1,500 <= Len < 2,000 | 17.71s | 22.36s | 14.93s |
| 2,000 <= Len < 3,000 | 26.97s | 36.68s | 20.99s |
| 3,000 <= Len < 5,000 | 45.56s | 58.42s | 35.82s |
| 5,000 <= Len < 8,000 | 56.57s | 58.17s | 55.55s |
| 8,000 <= Len < 10,000 | 57.76s | 58.86s | 56.66s |
| 10,000 <= Len | 66.49s | 76.80s | 58.42s |
| Protein Seq Len Range | Average Time | Maximum Time | Minimum Time |
|---|---|---|---|
| 300 <= Len < 500 | 1.89s | 2.55s | 1.10s |
| 500 <= Len < 800 | 2.68s | 3.44s | 2.13s |
| 800 <= Len < 1,000 | 3.45s | 4.25s | 2.65s |
| 1,000 <= Len < 1,500 | 4.27s | 5.90s | 3.54s |
| 1,500 <= Len < 2,000 | 5.81s | 7.44s | 4.76s |
| 2,000 <= Len < 3,000 | 8.14s | 10.74s | 6.37s |
| 3,000 <= Len < 5,000 | 13.25s | 17.69s | 10.06s |
| 5,000 <= Len < 8,000 | 17.03s | 18.20s | 15.98s |
| 8,000 <= Len < 10,000 | 17.90s | 18.99s | 16.92s |
| 10,000 <= Len | 25.90s | 35.02s | 18.66s |
-
Viral RdRP(Positive: 5,979)
The positive sequence fasta file is in
data/rdrp/all_dataset_positive.fasta.zip
all_dataset_positive.fasta.zip
or LucaProt Figshare -
Non-viral RdRP(Negative: 229434)
The negative sequence fasta file is in
dataset/rdrp/all_dataset_negative.fasta.zip
including:- other proteins of the virus
- other protein domains of the virus
- non-viral proteins
All structural embedding files of the dataset for model building are available at: embs
All structural embedding files of the prediction data for opening are in the process(because of the amount of data).
All 3D-structure PDB files of the model building dataset and predicted data for opening are in the process (because of the amount of data).
-
structure vocab
This vocab file isstruct_vocab/rdrp_40_extend/protein/binary_class/struct_vocab.txt
struct_vocab.txt -
subword-level vocab
The size of the vocab of sequence we use is 20,000.
This vocab file isvocab/rdrp_40_extend/protein/binary_class/subword_vocab_20000.txt
subword_vocab_20000.txt -
char-level vocab
This vocab file isvocab/rdrp_40_extend/protein/binary_class/vocab.txt
vocab.txt
Viral RdRP identification is a binary-class classification task, including positive and negative classes, using 0 and 1 to represent a negative and positive sample, respectively.
The label list file is dataset/rdrp_40_extend/protein/binary_class/label.txt
label.txt
We constructed a data set with 235,413 samples for model building, which included 5,979 positive samples of known viral RdRPs (i.e. the well-curated RdRP database described in the previous section of Methods), and 229,434 (to maintain a 1:40 ratio for viral RdRP and non-virus RdRPs) negative samples of confirmed non-virus RdRPs. And the non-virus RdRPs contained proteins from Eukaryota DNA dependent RNA polymerase (Eu DdRP, N=1,184), Eukaryota RNA dependent RNA polymerase (Eu RdRP, N=2,233), Reverse Transcriptase (RT, N=48,490), proteins obtained from DNA viruses (N=1,533), non-RdRP proteins obtained from RNA viruses (N=1,574), and a wide array of cellular proteins from different functional categories (N=174,420). We randomly divided the dataset into training, validation, and testing sets with a ratio of 8.5:1:1, which were used for model fitting, model finalization (based on the best F1-score training iteration), and performance reporting (including accuracy, precision, recall, F1-score, and Area under the ROC Curve (AUC)), respectively.
-
Entire Dataset
This file isdataset/rdrp/all_dataset_with_pdb_emb.csv.zip
all_dataset_with_pdb_emb.csv.zip
or LucaProt Figshare -
Training set
This file copy todataset/rdrp_40_extend/protein/binary_class/train_with_pdb_emb.csv
train_with_pdb_emb.csv
or LucaProt Figshare -
Validation set
This file copy todataset/rdrp_40_extend/protein/binary_class/dev_with_pdb_emb.csv
dev_with_pdb_emb.csv
or LucaProt Figshare -
Testing set
This file copy todataset/rdrp_40_extend/protein/binary_class/test_with_pdb_emb.csv
test_with_pdb_emb.csv
or LucaProt Figshare
One row in all the above files represents one sample. All three files consist of 9 columns, including prot_id, seq, seq_len, pdb_filename, ptm, mean_plddt, emb_filename, label, and source. The details of these columns are as follows:
- prot_id
the protein id - seq
the amino acid(aa) sequence - seq_len
the length of the protein sequence. - pdb_filename
The PDB filenames of 3D-structure are predicted by the calculation model or obtained by experiments. - ptm
the pTM of the predicted 3D-structure. - mean_plddt
the mean pLDDT of the predicted 3D-structure. - emb_filename
The filename of the embedding matrix or vector of protein structure.
Note: the embedding matrics of the dataset need to prepare in advance. - label
the sample label, 0 or 1 for binary-class classification, [0, 1, ..., N-1] for multi-class classification, a list of [0, 1, ..., N-1] for multi-label classification. - source
optional, the sample source (such as RdRP, RT, DdRP, non-virus RdRP, and Other).
Note: if using strategy one in structure encoder, the pdb_filename, the ptm, and the mean_plddt can be null.
You can use this project to train models for other tasks, not just the viral RdRP identification tasks.
-
binary-class classification
The label is 0 or 1 for binary-class classification, such as viral RdRP identification. -
multi-class classification
The label is 0~N-1 for multi-class classification, such as the species prediction for proteins. -
multi-label classification
The labels form a list of 0~N-1 for multi-label classification, such as Gene Ontology annotation for proteins.
The script structure_from_esm_v1.py is in the directory "src/protein_structure", and it use ESMFold (esmfold_v1) to predict 3D-Structure of protein.
cd LucaProt/src/protein_structure/
export CUDA_VISIBLE_DEVICES=0
python structure_from_esm_v1.py \
-i data/rdrp/rdrp.fasta \
-o pdbs/rdrp/ \
--num-recycles 4 \
--truncation_seq_length 4096 \
--chunk-size 64 \
--cpu-offload \
--batch_size 1
Parameters:
-
-i (input filepaths)
- fasta filepath
- csv filepath
the first row is the header
column 0: protein_id
column 1: sequence - mutil filepaths
comma-concatenation
-
-o (save dirpath)
The dir path of saving the predicted 3D-structure data, each protein is stored in a PDB file, and each PDB file is named as "protein_" + an auto-increment id + ".pdb", such as "protein_1.pdb".
The mapping between protein ids and auto-increment ids is stored in the file "result_info.csv" (including: "index", "protein_id(uuid)", "seq_len", "ptm", "mean_plddt") in this dir path.
For failed samples(CUDA out of memory), this script will save their protein ids in the "uncompleted.txt", and you can reduce the value of "truncation_seq_length" and add "--try_failure" for retry. -
--batch_size
the batch size of running, default: 1. -
--truncation_seq_length
truncate sequences longer than the given value, recommended values: 4096, 2048, 1984, 1792, 1536, 1280, 1152, 1022. -
--num-recycles
number of recycles to run. -
--chunk-size
chunks axial attention computation to reduce memory usage from O(L^2) to O(L), recommended values: 128, 64, 32. -
--try_failure
retry the failed samples when reducing the "truncation_seq_length" value.
cd LucaProt/src/protein_structure/
export CUDA_VISIBLE_DEVICES=0
python structure_from_esm_v1.py \
-name protein_id1,protein_id2 \
-seq VGGLFDYYSVPIMT,LPDSWENKLLTDLILFAGSFVGSDTCGKLF \
-o pdbs/rdrp/ \
--num-recycles 4 \
--truncation_seq_length 4096 \
--chunk-size 64 \
--cpu-offload \
--batch_size 1
Parameters:
- -name
protein ids, comma-concatenation for multi proteins. - -seq
protein sequences, comma-concatenation for multi proteins.
The script embedding_from_esmfold.py is in "src/protein_structure", and it use ESMFold (esm2_t36_3B_UR50D) to predict protein structural embedding matrices or vectors.
cd LucaProt/src/protein_structure/
export CUDA_VISIBLE_DEVICES=0
python embedding_from_esmfold.py \
--model_name esm2_t36_3B_UR50D \
--file data/rdrp.fasta \
--output_dir emb/rdrp/ \
--include per_tok contacts bos \
--truncation_seq_length 4094
Parameters:
-
--model_name
the model name, default: "esm2_t36_3B_UR50D" -
-i/--file (input filepath)
- fasta filepath
- csv filepath
the first row is the header
column 0: protein_id
column 1: sequence
-
-o/--output_dir (save dirpath)
The dir path of saving the predicted structural embedding data, each protein is stored in a pickle file, and each embedding file is named as "embedding_" + auto-increment id + ".pt", such as "embedding_1.pt".
The mapping between protein ids and auto-increment ids is stored in the file "{}_embed_fasta_id_2_idx.csv"(including: "index", "protein_id(uuid)") in this dir path.
For failed samples(CUDA out of memory), this script will save their protein ids in the "{}_embed_uncompleted.txt", and you can reduce the "truncation_seq_length" value and add "--try_failure" for retry. -
--truncation_seq_length
truncate sequences longer than the given value. Recommended values: 4094, 2046, 1982, 1790, 1534, 1278, 1150, 1022. -
--include
The embedding matrix or vector type of the predicted structural embedding data, including per_tok, mean, contacts, and bos.- per_tok includes the full sequence, with an embedding per amino acid (seq_len x hidden_dim).
- mean includes the embeddings averaged over the full sequence, per layer.
- bos includes the embeddings from the beginning-of-sequence token.
- contacts includes the attention value between two amino acids of the the full sequence.
Reference:https://github.com/facebookresearch/esm [Compute embeddings in bulk from FASTA]
cd LucaProt/src/protein_structure/
export CUDA_VISIBLE_DEVICES=0
python embedding_from_esmfold.py \
--model_name esm2_t36_3B_UR50D \
-name protein_id1,protein_id2 \
-seq VGGLFDYYSVPIMT,LPDSWENKLLTDLILFAGSFVGSDTCGKLF \
--output_dir embs/rdrp/test/ \
--include per_tok contacts bos \
--truncation_seq_length 4094
Parameters:
-
-name
protein ids, comma-concatenation for multi proteins. -
-seq
protein sequences, comma-concatenation for multi proteins.
Construct your dataset and randomly divide the dataset into training, validation, and testing sets with a specified ratio, and save the three sets in dataset/${dataset_name}/${dataset_type}/${task_type}, including train_.csv, dev_.csv, test_*.csv.
The file format can be .csv (must include the header ) or .txt (does not need to have the header).
Each file line is a sample containing 9 columns, including prot_id, seq, seq_len, pdb_filename, ptm, mean_plddt, emb_filename, label, and source.
Colunm seq is the sequence, Colunm pdb_filename is the saved PDB filename for structure encoder strategy 2, Colunm ptm and Column mean_plddt are optional, which are obtained from the 3D-Structure computed model, Colunm emb_filename is the saved embedding filename for structure encoder strategy 1, Column label is the sample class(a single value or a list value of label index or label name). Column source is the sample source (optional).
For example:
like_YP_009351861.1_Menghai_flavivirus,MEQNG...,3416,,,,embedding_21449.pt,1,rdrp
Note: if your dataset takes too much space to load into memory at once,
use "src/data_process/data_preprocess_into_tfrecords_for_rdrp.py" to convert the dataset into "tfrecords". And create an index file: python -m tfrecord.tools.tfrecord2idx xxxx.tfrecords xxxx.index
-
run.py
the main script for model building. -
Parameters
- data_dir: path, the dataset dirpath
- filename_pattern: the dataset filename pattern, such as "{}_with_pdb_emb.csv", including train_with_pdb_emb.csv, dev_with_pdb_emb.csv, and test_with_pdb_emb.csv in ${data_dir}
- separate_file: store_true, load the entire dataset into memory, the names of the pdb and embedding files are listed in the train/dev/test.csv, and need to load them.
- tfrecords: store_true, whether the dataset is in the tfrecords, when true, only the specified number of samples(${shuffle_queue_size}) are loaded into memory at once. The tfrecords must consist of "${data_dir}/tfrecords/train/xxx.tfrecords", "${data_dir}/tfrecords/dev/xxx.tfrecords" and "${data_dir}/tfrecords/test/xxx.tfrecords". "xxx.tfrecords" is one of 01-of-01.tfrecords(only including sequence), 01-of-01_emb.records (including sequence and structural embedding), and 01-of-01_pdb_emb.records (including sequence, 3D-structure contact map, and structural embedding).
- shuffle_queue_size: int, how many samples are loaded into memory at once, default: 5000.
- dataset_name: str, your dataset name, such as "rdrp_40_extend"
- dataset_type: str, your dataset type, such as "protein"
- task_type: choices=["multi_label", "multi_class", "binary_class"], your task type, such as "binary_class"
- model_type: choices=["sequence", "structure", "embedding", "sefn", "ssfn"], they represent only the sequence for input, only the 3D-structure contact map for input, only the structural embedding for input, the sequence and the structural embedding for input, and the sequence and the 3D-structure contact map for input, respectively
- subword: store_true, whether to process for sequence at the subword level.
- codes_file: path, subword codes filepath when using subword, such as "../subword/rdrp/protein_codes_rdrp_20000.txt"
- label_type: str, the label type name, such as "rdrp"
- label_filepath: path, the label list filepath
- cmap_type: choices=["C_alpha", "C_bert"], the calculation type of 3D-structure contact map
- cmap_thresh: the distance threshold (Unit: Angstrom) in contact map calculation. Two amino acids are linked if the distance between them is equal to and less than the threshold, default: 10.0.
- output_dir: path, the output dirpath
- log_dir: path, the logger savepath
- tb_log_dir: path, the save path of metric evaluation records in model training, the tensorboardX can be used to show these metrics.
- config_path: path, the configuration filepath of the model.
- seq_vocab_path: path, the vocab filepath of sequence tokenizer
- struct_vocab_path: path, the vocab filepath of 3D-structure node (Structural Encoder Strategy 2)
- seq_pooling_type: choices=["none", "max", "value_attention"], the sequence representation matrix pooling type, "none" represents that [CLS] vector is used.
- struct_pooling_type: choices=["max", "value_attention"], the 3D-structure representation matrix pooling type.
- embedding_pooling_type: choices=["none", "max", "value_attention"], the structural embedding representation matrix pooling type, "none" represents that [CLS] vector is used.
- evaluate_during_training: store_true, whether to evaluate the validation set and the testing set during training.
- do_eval: store_true, whether to use the best saved model to evaluate the validation set.
- do_predict: store_true, whether to use the best saved model to evaluate the testing set.
- do_lower_case: store_true, whether to lowercase the input when tokenizing.
- per_gpu_train_batch_size: int, batch size per GPU/CPU for training, default: 16
- per_gpu_eval_batch_size: int, batch size per GPU/CPU for evaluation, default: 16
- gradient_accumulation_steps: int, number of updates steps to accumulate before performing a backward/update pass, default: 1.
- learning_rate: float, the initial learning rate for Adam, default: 1e-4.
- num_train_epochs: int, the total number of training epochs to perform, default: 50.
- logging_steps: log every X updates steps, default: 1000.
- loss_type: choices=["focal_loss", "bce", "multilabel_cce", "asl", "cce"], loss-function type of model training, default: "bce".
- max_metric_type: choices=["acc", "jaccard", "prec", "recall", "f1", "fmax", "roc_auc", "pr_auc"], which metric is used for model finalization, default: "f1".
- pos_weight: float, positive samples weight for "bce".
- focal_loss_alpha: float, alpha for focal loss, default: 0.7.
- focal_loss_gamma: float, gamma for focal loss, default:2.0.
- focal_loss_reduce: store_true, "mean" for one sample when in multi-label classification, default:"sum".
- asl_gamma_neg: float, negative gamma for asl, default: 4.0.
- asl_gamma_pos: float, positive gamma for asl, default: 1.0.
- seq_max_length: int, the length of input sequence more than max length will be truncated, shorter will be padded, default: 2048.
- struct_max_length: int, the length of input contact map more than max length will be truncated, shorter will be padded., default: 2048.
- trunc_type: choices=["left", "right"], the truncate type for whole input sequence, default: "right".
- no_position_embeddings: store_true, whether not use position embedding for the sequence.
- no_token_type_embeddings: store_true, whether not use token type embedding for the sequence.
- embedding_input_size: int, the dim of the structural embedding vector/matrix, default: 2560, {"esm2_t30_150M_UR50D": 640, "esm2_t33_650M_UR50D": 1280, "esm2_t36_3B_UR50D": 2560, "esm2_t48_15B_UR50D": 5120}.
- embedding_type: choices=[None, "contacts", "bos", "matrix"], the type of the structural embedding info, default: "matrix".
- embedding_max_length: int, the length of input embedding matrix more than max length will be truncated, shorter will be padded, default: 2048.
- save_all: store_true, the model for each evaluation is saved.
- delete_old: store_true, only save the best metric (${max_metric_type}) model of all evaluation on testing set during training.
-
Training
#!/bin/bash export CUDA_VISIBLE_DEVICES=0 DATASET_NAME="rdrp_40_extend" DATASET_TYPE="protein" TASK_TYPE="binary_class" # sequence + structural embeddding MODEL_TYPE="sefn" CONFIG_NAME="sefn_config.json" INPUT_MODE="single" LABEL_TYPE="rdrp" embedding_input_size=2560 embedding_type="matrix" SEQ_MAX_LENGTH="2048" embedding_max_length="2048" TRUNCT_TYPE="right" # none, max, value_attention SEQ_POOLING_TYPE="value_attention" # max, value_attention embedding_pooling_type="value_attention" VOCAB_NAME="subword_vocab_20000.txt" SUBWORD_CODES_NAME="protein_codes_rdrp_20000.txt" MAX_METRIC_TYPE="f1" time_str=$(date "+%Y%m%d%H%M%S") python run.py \ --data_dir ../dataset/$DATASET_NAME/$DATASET_TYPE/$TASK_TYPE \ --tfrecords \ --filename_pattern {}_with_pdb_emb.csv \ --dataset_name $DATASET_NAME \ --dataset_type $DATASET_TYPE \ --task_type $TASK_TYPE \ --model_type $MODEL_TYPE \ --subword \ --codes_file ../subword/$DATASET_NAME/$DATASET_TYPE/$TASK_TYPE/$SUBWORD_CODES_NAME\ --input_mode $INPUT_MODE \ --label_type $LABEL_TYPE \ --label_filepath ../dataset/$DATASET_NAME/$DATASET_TYPE/$TASK_TYPE/label.txt \ --output_dir ../models/$DATASET_NAME/$DATASET_TYPE/$TASK_TYPE/$MODEL_TYPE/$time_str \ --log_dir ../logs/$DATASET_NAME/$DATASET_TYPE/$TASK_TYPE/$MODEL_TYPE/$time_str \ --tb_log_dir ../tb-logs/$DATASET_NAME/$DATASET_TYPE/$TASK_TYPE/$MODEL_TYPE/$time_str \ --config_path ../config/$DATASET_NAME/$DATASET_TYPE/$TASK_TYPE/$CONFIG_NAME \ --seq_vocab_path ../vocab/$DATASET_NAME/$DATASET_TYPE/$TASK_TYPE/$VOCAB_NAME\ --seq_pooling_type $SEQ_POOLING_TYPE \ --embedding_pooling_type $embedding_pooling_type \ --do_train \ --do_eval \ --do_predict \ --evaluate_during_training \ --per_gpu_train_batch_size=16 \ --per_gpu_eval_batch_size=16 \ --gradient_accumulation_steps=1 \ --learning_rate=1e-4 \ --num_train_epochs=50 \ --logging_steps=1000 \ --save_steps=1000 \ --overwrite_output_dir \ --sigmoid \ --loss_type bce \ --max_metric_type $MAX_METRIC_TYPE \ --seq_max_length=$SEQ_MAX_LENGTH \ --embedding_max_length=$embedding_max_length \ --trunc_type=$TRUNCT_TYPE \ --no_token_type_embeddings \ --embedding_input_size $embedding_input_size\ --embedding_type $embedding_type \ --shuffle_queue_size 10000 \ --save_all
-
Configuration file
The configuration files of all methods is in "config/rdrp_40_extend/protein/binary_class/".
If training your model, please put the configuration file in "config/${dataset_name}/${dataset_type}/${task_type}/" -
Value meaning in configuration file
referring to "src/SSFN/README.md" -
Baselines
-
LGBM (using the embedding vector: <bos> as the input)
cd src/baselines/ sh run_lgbm.sh -
XGBoost (using the embedding vector: <bos> as the input)
cd src/baselines/ sh run_xgb.sh -
DNN (using the embedding vector: <bos> as the input)
cd src/baselines/ sh run_dnn.shOr:
cd src/training run_subword_rdrp_emb.sh -
Transoformer-Char Level (using the sequence as the input)
cd src/training sh run_char_rdrp_seq.sh -
Transoformer-Subword Level (using the sequence as the input)
cd src/training sh run_subword_rdrp_seq.sh -
DNN2 (VALP + DNN, using the embedding matrix as the input)
cd src/training run_subword_rdrp_emb_v2.sh
-
-
Ours
-
Ours (the sequence + the 3D-structure)
coming soon... -
Ours (the sequence + the embedding matrix)
cd src/training run_subword_rdrp_sefn.sh
-
The running information is saved in "logs/${dataset_name}/${dataset_type}/${task_type}/${model_type}/${time_str}/logs.txt".
The information includes the model configuration, model layers, running parameters, and evaluation information.
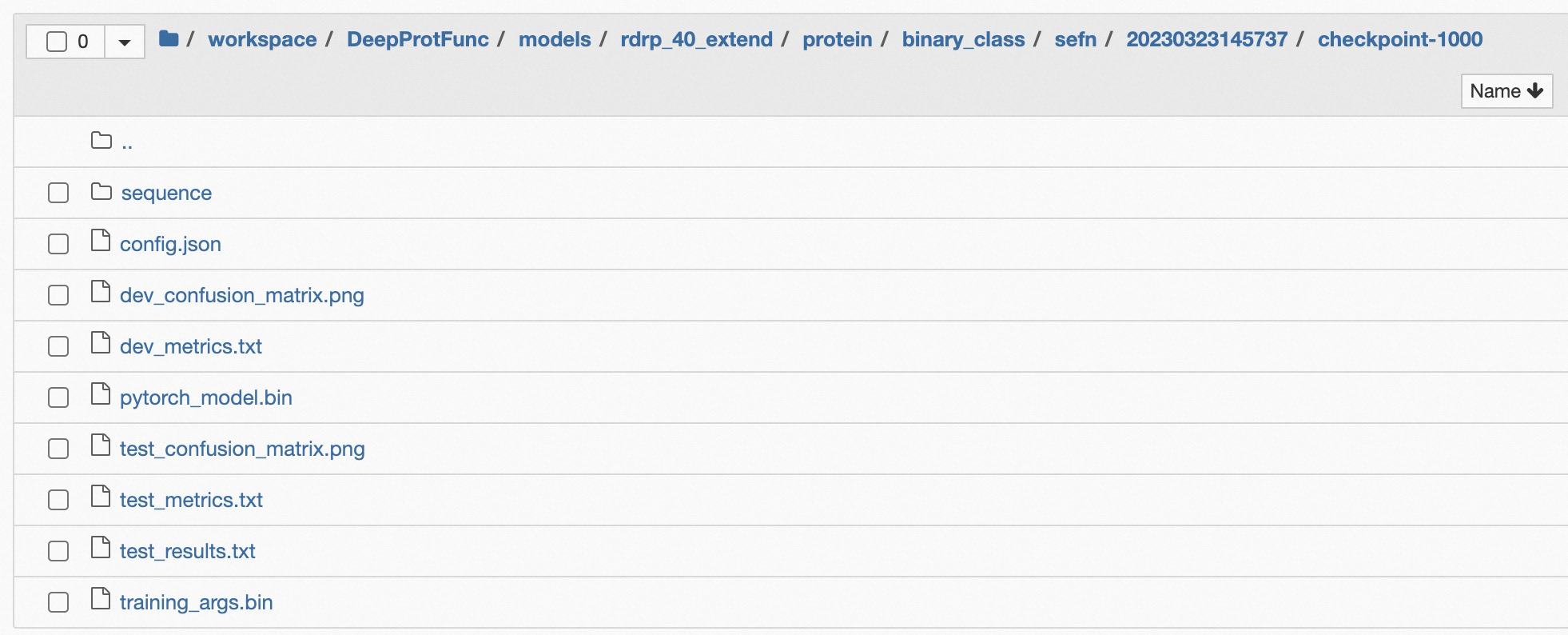
The checkpoints are saved in "models/${dataset_name}/${dataset_type}/${task_type}/${model_type}/${time_str}/checkpoint-${global_step}/", this directory includes "pytorch_model.bin", "config.json", "training_args.bin", and tokenizer information "sequence" or "strcut". The details are shown in Figure 2.
Figure 2: The File List in Checkpoint Dir Path
The metrics are recorded in "tb-logs/${dataset_name}/${dataset_type}/${task_type}/${model_type}/${time_str}/events.out.tfevents.xxxxx.xxxxx"
run: tensorboard --logdir=tb-logs/${dataset_name}/${dataset_type}/${task_type}/${model_type}/${time_str} --bind_all
The predicted results is saved in "predicts/${dataset_name}/${dataset_type}/${task_type}/${model_type}/${time_str}/checkpoint-${global_step}", including:
- pred_confusion_matrix.png
- pred_metrics.txt
- pred_result.csv
- seq_length_distribution.png
The details are shown in Figure 3.
Figure 3: The File List in Prediction Dir Path
Note: when using the saved model to predict, the "logs.txt" and the checkpoint dirpath will be used.
A conventional approach that clustered all proteins based on their sequence homology.
See ClstrSerch/README.md for details.
*.py in "src/data_preprocess"
*.py in "src/SSFN"
*.sh in "src/prediction"
including:
-
run_predict_from_file.sh
run prediction for many samples from a file, the structural embedding information prepared in advance. -
run_predict_one_sample.sh
run prediction for one sample from the input. -
run_predict_many_samples.sh
run prediction for many samples from the input.
We perform ablation studies on our model by removing specific module(sequence-specific and embedding-specific) one at a time to explore their relative importance.
-
run_predict_only_seq_from_file.sh
only using the sequence to predict and calculate metrics three positive testing datasets, three negative testing datasets, and our checked RdRPs by prediction SRA. -
run_predict_only_emb_from_file.sh
only using the structural embedding to predict and calculate metrics three positive testing datasets, three negative testing datasets, and our checked RdRPs by prediction SRA. -
run_predict_seq_emb_from_file.sh
using the sequentail info and the structural embedding to predict and calculate metrics three positive testing datasets, three negative testing datasets, and our checked RdRPs by prediction SRA.
*.py in "src/baselines", using the embedding vector as the input, including:
- DNN
- LGBM
- XGBoost
*.py in "src/deep_baselines", including:
CHEER: HierarCHical taxonomic classification for viral mEtagEnomic data via deep learning(2021). code: CHEER
VirHunter: A Deep Learning-Based Method for Detection of Novel RNA Viruses in Plant Sequencing Data(2022). code: VirHunter
Virtifier: a deep learning-based identifier for viral sequences from metagenomes(2022). code: Virtifier
RNN-VirSeeker: RNN-VirSeeker: A Deep Learning Method for Identification of Short Viral Sequences From Metagenomes. code: RNN-VirSeeker
-
run_deep_baselines.sh
the script to train deep baseline models. -
run_predict_deep_baselines.sh
use trained deep baseline models to predict three positive test datasets, three negative test datasets, and our checked RdRP datasets. -
run.py
the main script for training deep baseline models. -
statistics
the script to statistic the accuracy in three kinds of test datasets(positive, negative, our checked) after prediction by deep baselines.
*.py in "src/biotoolbox"
*.py in "src/common"
*.sh in "src/training"
*.sh in "src/prediction"
the raw data is in "data/".
the files of the dataset is in "dataset/${dataset_name}/${dataset_type}/${task_type}/".
the configuration file of all methods is in "config/${dataset_name}/${dataset_type}/${task_type}/".
some pictures is in "pics/".
the scripts of pictures ploting is in "src/plot".
the codes and results of Geo information Spider in "src/geo_map".
The open resources of our study ar includes 10 subdirectories: Known_RdRPs, Benchmark, Results, All_Contigs, All_Protein_Sequences, SG_predicted_protein_structure_supplementation/, Serratus, Self_Sequencing_Proteins, Self_Sequencing_Reads, and LucaProt.
Please refer to README.md or LucaProt Figshare .
LucaProt/ includes some resources related to LucaProt, including dataset for model building(dataset_for_model_building), dataset for_model evaluation(dataset_for_model_evaluation), and our trained model(logs/ and models/).
As mentioned above.
-
sequential info
1) train_with_pdb_emb.csv
copy toLucaProt/dataset/rdrp_40_extend/protein/binary_class/
or LucaProt Figshare2) dev_with_pdb_emb.csv
copy toLucaProt/dataset/rdrp_40_extend/protein/binary_class/
or LucaProt Figshare3) test_with_pdb_emb.csv
copy toLucaProt/dataset/rdrp_40_extend/protein/binary_class/
or LucaProt Figshare -
structural info
embs
copy toLucaProt/dataset/rdrp_40_extend/protein/binary_class/embs/ -
tfrcords
1) train
copy toLucaProt/dataset/rdrp_40_extend/protein/binary_class/tfrecords/train/2) dev
copy toLucaProt/dataset/rdrp_40_extend/protein/binary_class/tfrecords/dev/3) test
copy toLucaProt/dataset/rdrp_40_extend/protein/binary_class/tfrecords/test/
-
7 Positive Testing Datasets
1) Wolf et al., 2020 NM
Reference: Doubling of the known set of RNA viruses by metagenomic analysis of an aquatic virome.
or LucaProt Figshare2) Edgar et al., 2022 Nature
Reference: Petabase-scale sequence alignment catalyses viral discovery.
or LucaProt Figshare3) Zayed et al, 2022 Science
Reference: Cryptic and abundant marine viruses at the evolutionary origins of Earth's RNA virome.
or LucaProt Figshare4) Neri et al., 2022 Cell
Reference: Expansion of the global RNA virome reveals diverse clades of bacteriophages.
or LucaProt Figshare5) Chen et al., 2022 NM
Reference: RNA viromes from terrestrial sites across China expand environmental viral diversity.
or LucaProt Figshare6) Olendraite et al., 2023 MBE
Reference: Identification of RNA Virus-Derived RdRp Sequences in Publicly Available Transcriptomic Data Sets.
or LucaProt Figshare7) This Study
Reference: Artificial intelligence redefines RNA virus discovery.
or LucaProt Figshare -
4 Negative Testing Datasets
1) RT
or LucaProt Figshare2) Eu DdRP
or LucaProt Figshare
- Our Checked RdRP Dataset (Our Results)
-
sequential info
ours_checked_rdrp_final.csv
or LucaProt Figshare -
structural info
embs -
PDB
All 3D-structure PDB files of our predicted results for opening are in the process.
-
- Our Sampled Dataset
The trained model for RdRP identification is available at:
Notice: these files were already downloaded in this GitHub project, so you don't need to download them.
-
logs
logs
copy toLucaProt/logs/
or LucaProt Figshare -
models
models
copy toLucaProt/models/
or LucaProt Figshare
LucaTeam:
Yong He, Zhaorong Li, Xin Hou, Mang Shi, Pan Fang
FTP: The all data of LucaProt is available at the website: Open Resources
Figshare: https://doi.org/10.6084/m9.figshare.26298802.v13
the pre-print version:
@article { lucaprot,
author = {Xin Hou and Yong He and Pan Fang and Shi-Qiang Mei and Zan Xu and Wei-Chen Wu and Jun-Hua Tian and Shun Zhang and Zhen-Yu Zeng and Qin-Yu Gou and Gen-Yang Xin and Shi-Jia Le and Yin-Yue Xia and Yu-Lan Zhou and Feng-Ming Hui and Yuan-Fei Pan and John-Sebastian Eden and Zhao-Hui Yang and Chong Han and Yue-Long Shu and Deyin Guo and Jun Li and Edward C Holmes and Zhao-Rong Li and Mang Shi},
title = {Artificial intelligence redefines RNA virus discovery},
elocation-id = {2023.04.18.537342},
year = {2023},
doi = {10.1101/2023.04.18.537342},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2023/04/18/2023.04.18.537342 },
eprint = {https://www.biorxiv.org/content/early/2023/04/18/2023.04.18.537342.full.pdf },
journal = {bioRxiv}
}