This module should preferably be used to test unimodal distributions.
Let
Let us partition the samples
so that each bins is characterised by an observed numerosity
As the best practice suggests, should the i-th class have
It is widely known that, under
where
In order to accomplish the test we need to set a significativity level thereof:
First we need to import the necessary modules.
from pygof.test import chi2test
from pygof.binning import re_bin
from pygof.util import random_variable, inspect_sample
import scipy.stats
import numpy as np
np.random.seed(1) # for repeatabilityIf we just wish to include the package without installing it, we need to include this as well:
import sys
sys.path.append("../src/")Assume that our sample has 5000 items, grouped in 50 bins:
sample_size = 5000
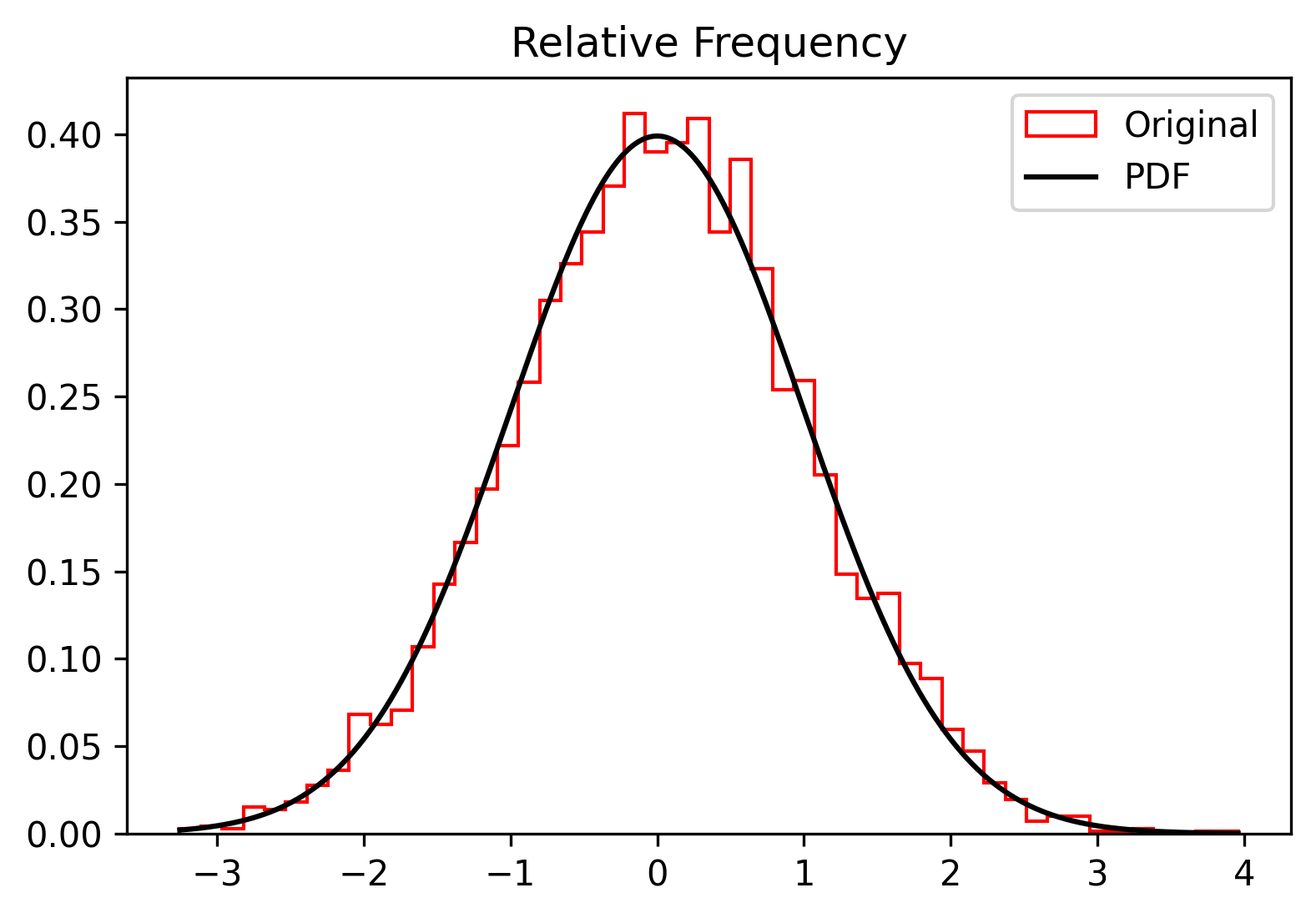
num_bins = 50We define a standardised Normal random variable whereby we generate a sample onto which we shall perform the scipy functions to define the random variable and we then generate a sample. Since we already known the distribution governing the sample we should be getting just a confirmation! Needless to say, this sample can be replaced with a real sample of a case study.
rv = random_variable(scipy.stats.norm, loc=0, scale=1)
sample = rv.rvs(size=sample_size)As mentioned earlier, each bin must contain at least 5 items to conduct a meaninful th=5, and rebin the sample by:
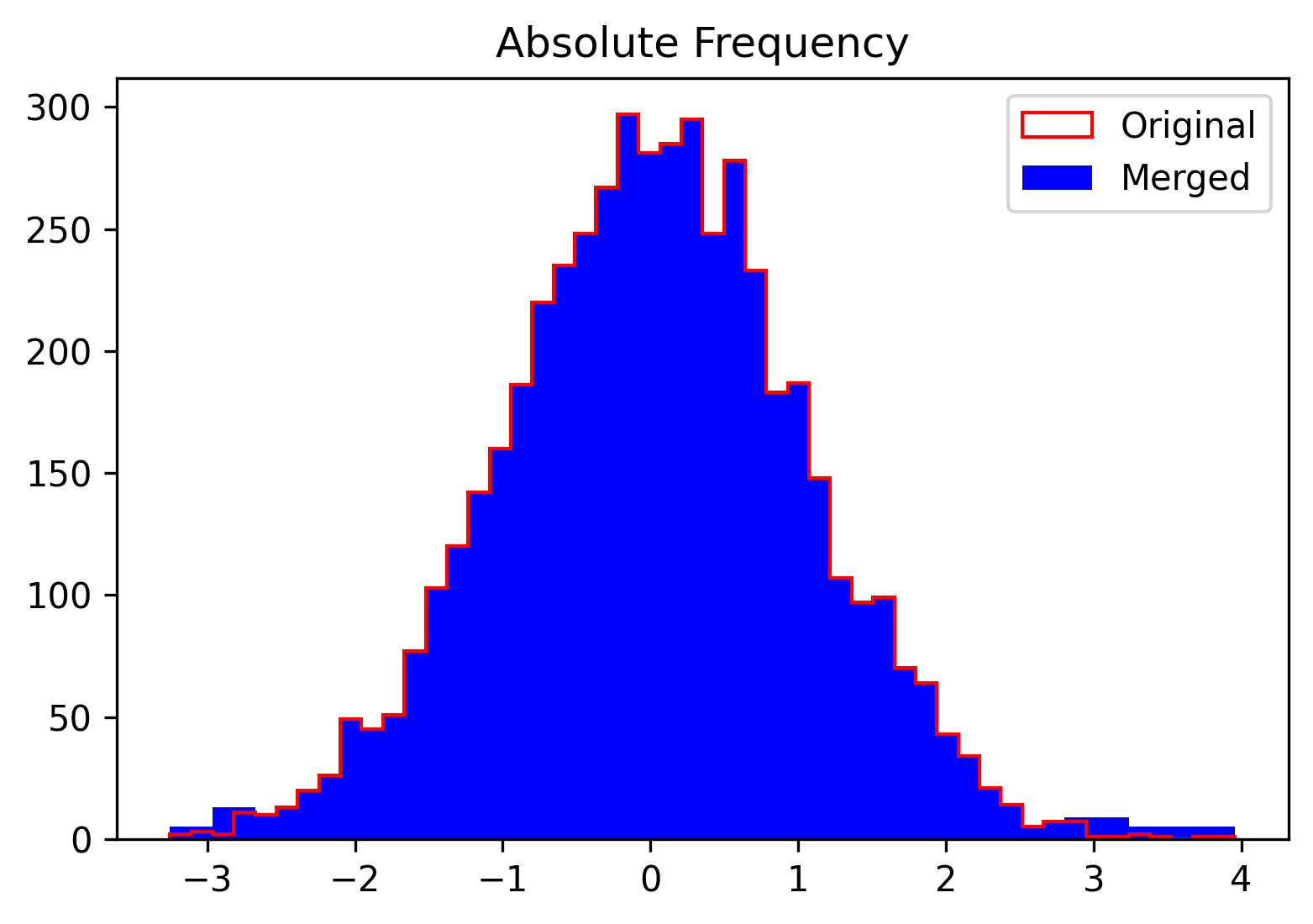
merged_counts, merged_edges, merged_edges_plot = re_bin(sample, n_bins=num_bins, th=th)where merged_edges_plot are returned only for visualisation purposes. Finally the
chi2test(merged_counts, merged_edges, rv, signif=10, est_params=True)provides the results we anticipated:
------------------------------------------------------------
Number of samples: 5000
Number of bins: 42
Number of Degrees of Freedom (DoFs): 39
Number of Estimated Parameters: 2
Significativity = 10.00%
Chi2 from data = 36.45
Chi2 from function = 50.66
Chi2 test PASSED (data < function)
------------------------------------------------------------We can also inspect the sample:
inspect_sample(sample, n_bins=num_bins, rv=rv, density=True)and the results:
inspect_sample(sample, n_bins=num_bins, rv=rv, density=False,
new_counts=merged_counts, new_edges=merged_edges_plot)